biopipen 0.21.0__py3-none-any.whl → 0.34.26__py3-none-any.whl
This diff represents the content of publicly available package versions that have been released to one of the supported registries. The information contained in this diff is provided for informational purposes only and reflects changes between package versions as they appear in their respective public registries.
- biopipen/__init__.py +1 -1
- biopipen/core/config.toml +28 -0
- biopipen/core/filters.py +79 -4
- biopipen/core/proc.py +12 -3
- biopipen/core/testing.py +75 -3
- biopipen/ns/bam.py +148 -6
- biopipen/ns/bed.py +75 -0
- biopipen/ns/cellranger.py +186 -0
- biopipen/ns/cellranger_pipeline.py +126 -0
- biopipen/ns/cnv.py +19 -3
- biopipen/ns/cnvkit.py +1 -1
- biopipen/ns/cnvkit_pipeline.py +20 -12
- biopipen/ns/delim.py +34 -35
- biopipen/ns/gene.py +68 -23
- biopipen/ns/gsea.py +63 -37
- biopipen/ns/misc.py +39 -14
- biopipen/ns/plot.py +304 -1
- biopipen/ns/protein.py +183 -0
- biopipen/ns/regulatory.py +290 -0
- biopipen/ns/rnaseq.py +142 -5
- biopipen/ns/scrna.py +2053 -473
- biopipen/ns/scrna_metabolic_landscape.py +228 -382
- biopipen/ns/snp.py +659 -0
- biopipen/ns/stats.py +484 -0
- biopipen/ns/tcr.py +683 -98
- biopipen/ns/vcf.py +236 -2
- biopipen/ns/web.py +97 -6
- biopipen/reports/bam/CNVpytor.svelte +4 -9
- biopipen/reports/cellranger/CellRangerCount.svelte +18 -0
- biopipen/reports/cellranger/CellRangerSummary.svelte +16 -0
- biopipen/reports/cellranger/CellRangerVdj.svelte +18 -0
- biopipen/reports/cnvkit/CNVkitDiagram.svelte +1 -1
- biopipen/reports/cnvkit/CNVkitHeatmap.svelte +1 -1
- biopipen/reports/cnvkit/CNVkitScatter.svelte +1 -1
- biopipen/reports/common.svelte +15 -0
- biopipen/reports/protein/ProdigySummary.svelte +16 -0
- biopipen/reports/scrna/CellsDistribution.svelte +4 -39
- biopipen/reports/scrna/DimPlots.svelte +1 -1
- biopipen/reports/scrna/MarkersFinder.svelte +6 -126
- biopipen/reports/scrna/MetaMarkers.svelte +3 -75
- biopipen/reports/scrna/RadarPlots.svelte +4 -20
- biopipen/reports/scrna_metabolic_landscape/MetabolicFeatures.svelte +61 -22
- biopipen/reports/scrna_metabolic_landscape/MetabolicPathwayActivity.svelte +88 -82
- biopipen/reports/scrna_metabolic_landscape/MetabolicPathwayHeterogeneity.svelte +70 -10
- biopipen/reports/snp/PlinkCallRate.svelte +24 -0
- biopipen/reports/snp/PlinkFreq.svelte +18 -0
- biopipen/reports/snp/PlinkHWE.svelte +18 -0
- biopipen/reports/snp/PlinkHet.svelte +18 -0
- biopipen/reports/snp/PlinkIBD.svelte +18 -0
- biopipen/reports/tcr/CDR3AAPhyschem.svelte +19 -66
- biopipen/reports/tcr/ClonalStats.svelte +16 -0
- biopipen/reports/tcr/CloneResidency.svelte +3 -93
- biopipen/reports/tcr/Immunarch.svelte +4 -155
- biopipen/reports/tcr/TCRClusterStats.svelte +3 -45
- biopipen/reports/tcr/TESSA.svelte +11 -28
- biopipen/reports/utils/misc.liq +22 -7
- biopipen/scripts/bam/BamMerge.py +11 -15
- biopipen/scripts/bam/BamSampling.py +90 -0
- biopipen/scripts/bam/BamSort.py +141 -0
- biopipen/scripts/bam/BamSplitChroms.py +10 -10
- biopipen/scripts/bam/BamSubsetByBed.py +38 -0
- biopipen/scripts/bam/CNAClinic.R +41 -5
- biopipen/scripts/bam/CNVpytor.py +153 -54
- biopipen/scripts/bam/ControlFREEC.py +13 -14
- biopipen/scripts/bam/SamtoolsView.py +33 -0
- biopipen/scripts/bed/Bed2Vcf.py +5 -5
- biopipen/scripts/bed/BedConsensus.py +5 -5
- biopipen/scripts/bed/BedLiftOver.sh +6 -4
- biopipen/scripts/bed/BedtoolsIntersect.py +54 -0
- biopipen/scripts/bed/BedtoolsMakeWindows.py +47 -0
- biopipen/scripts/bed/BedtoolsMerge.py +4 -4
- biopipen/scripts/cellranger/CellRangerCount.py +138 -0
- biopipen/scripts/cellranger/CellRangerSummary.R +181 -0
- biopipen/scripts/cellranger/CellRangerVdj.py +112 -0
- biopipen/scripts/cnv/AneuploidyScore.R +55 -20
- biopipen/scripts/cnv/AneuploidyScoreSummary.R +221 -163
- biopipen/scripts/cnv/TMADScore.R +25 -9
- biopipen/scripts/cnv/TMADScoreSummary.R +57 -86
- biopipen/scripts/cnvkit/CNVkitAccess.py +7 -6
- biopipen/scripts/cnvkit/CNVkitAutobin.py +26 -18
- biopipen/scripts/cnvkit/CNVkitBatch.py +6 -6
- biopipen/scripts/cnvkit/CNVkitCall.py +3 -3
- biopipen/scripts/cnvkit/CNVkitCoverage.py +4 -3
- biopipen/scripts/cnvkit/CNVkitDiagram.py +5 -5
- biopipen/scripts/cnvkit/CNVkitFix.py +3 -3
- biopipen/scripts/cnvkit/CNVkitGuessBaits.py +12 -8
- biopipen/scripts/cnvkit/CNVkitHeatmap.py +5 -5
- biopipen/scripts/cnvkit/CNVkitReference.py +6 -5
- biopipen/scripts/cnvkit/CNVkitScatter.py +5 -5
- biopipen/scripts/cnvkit/CNVkitSegment.py +5 -5
- biopipen/scripts/cnvkit/guess_baits.py +166 -93
- biopipen/scripts/delim/RowsBinder.R +1 -1
- biopipen/scripts/delim/SampleInfo.R +116 -118
- biopipen/scripts/gene/GeneNameConversion.R +67 -0
- biopipen/scripts/gene/GenePromoters.R +61 -0
- biopipen/scripts/gsea/Enrichr.R +5 -5
- biopipen/scripts/gsea/FGSEA.R +184 -50
- biopipen/scripts/gsea/GSEA.R +2 -2
- biopipen/scripts/gsea/PreRank.R +5 -5
- biopipen/scripts/misc/Config2File.py +2 -2
- biopipen/scripts/misc/Plot.R +80 -0
- biopipen/scripts/misc/Shell.sh +15 -0
- biopipen/scripts/misc/Str2File.py +2 -2
- biopipen/scripts/plot/Heatmap.R +3 -3
- biopipen/scripts/plot/Manhattan.R +147 -0
- biopipen/scripts/plot/QQPlot.R +146 -0
- biopipen/scripts/plot/ROC.R +88 -0
- biopipen/scripts/plot/Scatter.R +112 -0
- biopipen/scripts/plot/VennDiagram.R +5 -9
- biopipen/scripts/protein/MMCIF2PDB.py +33 -0
- biopipen/scripts/protein/PDB2Fasta.py +60 -0
- biopipen/scripts/protein/Prodigy.py +119 -0
- biopipen/scripts/protein/ProdigySummary.R +140 -0
- biopipen/scripts/protein/RMSD.py +178 -0
- biopipen/scripts/regulatory/MotifAffinityTest.R +102 -0
- biopipen/scripts/regulatory/MotifAffinityTest_AtSNP.R +127 -0
- biopipen/scripts/regulatory/MotifAffinityTest_MotifBreakR.R +104 -0
- biopipen/scripts/regulatory/MotifScan.py +159 -0
- biopipen/scripts/regulatory/VariantMotifPlot.R +78 -0
- biopipen/scripts/regulatory/motifs-common.R +324 -0
- biopipen/scripts/rnaseq/Simulation-ESCO.R +180 -0
- biopipen/scripts/rnaseq/Simulation-RUVcorr.R +45 -0
- biopipen/scripts/rnaseq/Simulation.R +21 -0
- biopipen/scripts/rnaseq/UnitConversion.R +325 -54
- biopipen/scripts/scrna/AnnData2Seurat.R +40 -0
- biopipen/scripts/scrna/CCPlotR-patch.R +161 -0
- biopipen/scripts/scrna/CellCellCommunication.py +150 -0
- biopipen/scripts/scrna/CellCellCommunicationPlots.R +93 -0
- biopipen/scripts/scrna/CellSNPLite.py +30 -0
- biopipen/scripts/scrna/CellTypeAnnotation-celltypist.R +185 -0
- biopipen/scripts/scrna/CellTypeAnnotation-direct.R +68 -31
- biopipen/scripts/scrna/CellTypeAnnotation-hitype.R +27 -22
- biopipen/scripts/scrna/CellTypeAnnotation-sccatch.R +28 -20
- biopipen/scripts/scrna/CellTypeAnnotation-sctype.R +48 -25
- biopipen/scripts/scrna/CellTypeAnnotation.R +37 -1
- biopipen/scripts/scrna/CellsDistribution.R +456 -167
- biopipen/scripts/scrna/DimPlots.R +1 -1
- biopipen/scripts/scrna/ExprImputation-alra.R +109 -0
- biopipen/scripts/scrna/ExprImputation-rmagic.R +256 -0
- biopipen/scripts/scrna/{ExprImpution-scimpute.R → ExprImputation-scimpute.R} +8 -5
- biopipen/scripts/scrna/ExprImputation.R +7 -0
- biopipen/scripts/scrna/LoomTo10X.R +51 -0
- biopipen/scripts/scrna/MQuad.py +25 -0
- biopipen/scripts/scrna/MarkersFinder.R +679 -400
- biopipen/scripts/scrna/MetaMarkers.R +265 -161
- biopipen/scripts/scrna/ModuleScoreCalculator.R +66 -11
- biopipen/scripts/scrna/PseudoBulkDEG.R +678 -0
- biopipen/scripts/scrna/RadarPlots.R +355 -134
- biopipen/scripts/scrna/ScFGSEA.R +298 -100
- biopipen/scripts/scrna/ScSimulation.R +65 -0
- biopipen/scripts/scrna/ScVelo.py +617 -0
- biopipen/scripts/scrna/Seurat2AnnData.R +7 -0
- biopipen/scripts/scrna/SeuratClusterStats-clustree.R +87 -0
- biopipen/scripts/scrna/SeuratClusterStats-dimplots.R +36 -30
- biopipen/scripts/scrna/SeuratClusterStats-features.R +138 -187
- biopipen/scripts/scrna/SeuratClusterStats-ngenes.R +81 -0
- biopipen/scripts/scrna/SeuratClusterStats-stats.R +78 -89
- biopipen/scripts/scrna/SeuratClusterStats.R +47 -10
- biopipen/scripts/scrna/SeuratClustering.R +36 -233
- biopipen/scripts/scrna/SeuratLoading.R +2 -2
- biopipen/scripts/scrna/SeuratMap2Ref.R +84 -113
- biopipen/scripts/scrna/SeuratMetadataMutater.R +16 -6
- biopipen/scripts/scrna/SeuratPreparing.R +223 -173
- biopipen/scripts/scrna/SeuratSubClustering.R +64 -0
- biopipen/scripts/scrna/SeuratTo10X.R +27 -0
- biopipen/scripts/scrna/Slingshot.R +65 -0
- biopipen/scripts/scrna/Subset10X.R +2 -2
- biopipen/scripts/scrna/TopExpressingGenes.R +169 -135
- biopipen/scripts/scrna/celltypist-wrapper.py +195 -0
- biopipen/scripts/scrna/scvelo_paga.py +313 -0
- biopipen/scripts/scrna/seurat_anndata_conversion.py +98 -0
- biopipen/scripts/scrna_metabolic_landscape/MetabolicFeatures.R +447 -82
- biopipen/scripts/scrna_metabolic_landscape/MetabolicPathwayActivity.R +348 -241
- biopipen/scripts/scrna_metabolic_landscape/MetabolicPathwayHeterogeneity.R +188 -166
- biopipen/scripts/snp/MatrixEQTL.R +217 -0
- biopipen/scripts/snp/Plink2GTMat.py +148 -0
- biopipen/scripts/snp/PlinkCallRate.R +199 -0
- biopipen/scripts/snp/PlinkFilter.py +100 -0
- biopipen/scripts/snp/PlinkFreq.R +291 -0
- biopipen/scripts/snp/PlinkFromVcf.py +81 -0
- biopipen/scripts/snp/PlinkHWE.R +85 -0
- biopipen/scripts/snp/PlinkHet.R +96 -0
- biopipen/scripts/snp/PlinkIBD.R +196 -0
- biopipen/scripts/snp/PlinkSimulation.py +124 -0
- biopipen/scripts/snp/PlinkUpdateName.py +124 -0
- biopipen/scripts/stats/ChowTest.R +146 -0
- biopipen/scripts/stats/DiffCoexpr.R +152 -0
- biopipen/scripts/stats/LiquidAssoc.R +135 -0
- biopipen/scripts/stats/Mediation.R +108 -0
- biopipen/scripts/stats/MetaPvalue.R +130 -0
- biopipen/scripts/stats/MetaPvalue1.R +74 -0
- biopipen/scripts/tcgamaf/Maf2Vcf.py +2 -2
- biopipen/scripts/tcgamaf/MafAddChr.py +2 -2
- biopipen/scripts/tcr/Attach2Seurat.R +3 -2
- biopipen/scripts/tcr/CDR3AAPhyschem.R +211 -143
- biopipen/scripts/tcr/CDR3Clustering.R +343 -0
- biopipen/scripts/tcr/ClonalStats.R +526 -0
- biopipen/scripts/tcr/CloneResidency.R +255 -131
- biopipen/scripts/tcr/CloneSizeQQPlot.R +4 -4
- biopipen/scripts/tcr/GIANA/GIANA.py +1356 -797
- biopipen/scripts/tcr/GIANA/GIANA4.py +1362 -789
- biopipen/scripts/tcr/GIANA/query.py +164 -162
- biopipen/scripts/tcr/Immunarch-basic.R +31 -9
- biopipen/scripts/tcr/Immunarch-clonality.R +25 -5
- biopipen/scripts/tcr/Immunarch-diversity.R +352 -134
- biopipen/scripts/tcr/Immunarch-geneusage.R +45 -5
- biopipen/scripts/tcr/Immunarch-kmer.R +68 -8
- biopipen/scripts/tcr/Immunarch-overlap.R +84 -4
- biopipen/scripts/tcr/Immunarch-spectratyping.R +35 -6
- biopipen/scripts/tcr/Immunarch-tracking.R +38 -6
- biopipen/scripts/tcr/Immunarch-vjjunc.R +165 -0
- biopipen/scripts/tcr/Immunarch.R +63 -11
- biopipen/scripts/tcr/Immunarch2VDJtools.R +2 -2
- biopipen/scripts/tcr/ImmunarchFilter.R +4 -4
- biopipen/scripts/tcr/ImmunarchLoading.R +38 -29
- biopipen/scripts/tcr/SampleDiversity.R +1 -1
- biopipen/scripts/tcr/ScRepCombiningExpression.R +40 -0
- biopipen/scripts/tcr/ScRepLoading.R +166 -0
- biopipen/scripts/tcr/TCRClusterStats.R +176 -22
- biopipen/scripts/tcr/TCRDock.py +110 -0
- biopipen/scripts/tcr/TESSA.R +102 -118
- biopipen/scripts/tcr/VJUsage.R +5 -5
- biopipen/scripts/tcr/immunarch-patched.R +142 -0
- biopipen/scripts/tcr/vdjtools-patch.sh +1 -1
- biopipen/scripts/vcf/BcftoolsAnnotate.py +91 -0
- biopipen/scripts/vcf/BcftoolsFilter.py +90 -0
- biopipen/scripts/vcf/BcftoolsMerge.py +31 -0
- biopipen/scripts/vcf/BcftoolsSort.py +113 -0
- biopipen/scripts/vcf/BcftoolsView.py +73 -0
- biopipen/scripts/vcf/TruvariBench.sh +14 -7
- biopipen/scripts/vcf/TruvariBenchSummary.R +16 -13
- biopipen/scripts/vcf/TruvariConsistency.R +1 -1
- biopipen/scripts/vcf/Vcf2Bed.py +2 -2
- biopipen/scripts/vcf/VcfAnno.py +11 -11
- biopipen/scripts/vcf/VcfDownSample.sh +22 -10
- biopipen/scripts/vcf/VcfFilter.py +5 -5
- biopipen/scripts/vcf/VcfFix.py +7 -7
- biopipen/scripts/vcf/VcfFix_utils.py +13 -4
- biopipen/scripts/vcf/VcfIndex.py +3 -3
- biopipen/scripts/vcf/VcfIntersect.py +3 -3
- biopipen/scripts/vcf/VcfLiftOver.sh +5 -0
- biopipen/scripts/vcf/VcfSplitSamples.py +4 -4
- biopipen/scripts/vcf/bcftools_utils.py +52 -0
- biopipen/scripts/web/Download.py +8 -4
- biopipen/scripts/web/DownloadList.py +5 -5
- biopipen/scripts/web/GCloudStorageDownloadBucket.py +82 -0
- biopipen/scripts/web/GCloudStorageDownloadFile.py +23 -0
- biopipen/scripts/web/gcloud_common.py +49 -0
- biopipen/utils/gene.py +108 -60
- biopipen/utils/misc.py +146 -20
- biopipen/utils/reference.py +64 -20
- biopipen/utils/reporter.py +177 -0
- biopipen/utils/vcf.py +1 -1
- biopipen-0.34.26.dist-info/METADATA +27 -0
- biopipen-0.34.26.dist-info/RECORD +292 -0
- {biopipen-0.21.0.dist-info → biopipen-0.34.26.dist-info}/WHEEL +1 -1
- {biopipen-0.21.0.dist-info → biopipen-0.34.26.dist-info}/entry_points.txt +6 -2
- biopipen/ns/bcftools.py +0 -111
- biopipen/ns/scrna_basic.py +0 -255
- biopipen/reports/delim/SampleInfo.svelte +0 -36
- biopipen/reports/scrna/GeneExpressionInvistigation.svelte +0 -32
- biopipen/reports/scrna/ScFGSEA.svelte +0 -35
- biopipen/reports/scrna/SeuratClusterStats.svelte +0 -82
- biopipen/reports/scrna/SeuratMap2Ref.svelte +0 -20
- biopipen/reports/scrna/SeuratPreparing.svelte +0 -38
- biopipen/reports/scrna/TopExpressingGenes.svelte +0 -55
- biopipen/reports/scrna_metabolic_landscape/MetabolicFeaturesIntraSubset.svelte +0 -31
- biopipen/reports/utils/gsea.liq +0 -110
- biopipen/scripts/bcftools/BcftoolsAnnotate.py +0 -42
- biopipen/scripts/bcftools/BcftoolsFilter.py +0 -79
- biopipen/scripts/bcftools/BcftoolsSort.py +0 -19
- biopipen/scripts/gene/GeneNameConversion.py +0 -66
- biopipen/scripts/scrna/ExprImpution-alra.R +0 -32
- biopipen/scripts/scrna/ExprImpution-rmagic.R +0 -29
- biopipen/scripts/scrna/ExprImpution.R +0 -7
- biopipen/scripts/scrna/GeneExpressionInvistigation.R +0 -132
- biopipen/scripts/scrna/Write10X.R +0 -11
- biopipen/scripts/scrna_metabolic_landscape/MetabolicFeaturesIntraSubset.R +0 -150
- biopipen/scripts/tcr/TCRClustering.R +0 -280
- biopipen/utils/common_docstrs.py +0 -61
- biopipen/utils/gene.R +0 -49
- biopipen/utils/gsea.R +0 -193
- biopipen/utils/io.R +0 -20
- biopipen/utils/misc.R +0 -114
- biopipen/utils/mutate_helpers.R +0 -433
- biopipen/utils/plot.R +0 -173
- biopipen/utils/rnaseq.R +0 -48
- biopipen/utils/single_cell.R +0 -115
- biopipen-0.21.0.dist-info/METADATA +0 -22
- biopipen-0.21.0.dist-info/RECORD +0 -218
biopipen/ns/stats.py
ADDED
|
@@ -0,0 +1,484 @@
|
|
|
1
|
+
"""Provides processes for statistics."""
|
|
2
|
+
|
|
3
|
+
from ..core.proc import Proc
|
|
4
|
+
from ..core.config import config
|
|
5
|
+
|
|
6
|
+
|
|
7
|
+
class ChowTest(Proc):
|
|
8
|
+
"""Massive Chow tests.
|
|
9
|
+
|
|
10
|
+
See Also https://en.wikipedia.org/wiki/Chow_test
|
|
11
|
+
|
|
12
|
+
Input:
|
|
13
|
+
infile: The input data file. The rows are samples and the columns are
|
|
14
|
+
features. It must be tab-delimited.
|
|
15
|
+
```
|
|
16
|
+
Sample F1 F2 F3 ... Fn
|
|
17
|
+
S1 1.2 3.4 5.6 7.8
|
|
18
|
+
S2 2.3 4.5 6.7 8.9
|
|
19
|
+
...
|
|
20
|
+
Sm 5.6 7.8 9.0 1.2
|
|
21
|
+
```
|
|
22
|
+
groupfile: The group file. The rows are the samples and the columns
|
|
23
|
+
are the groupings. It must be tab-delimited.
|
|
24
|
+
```
|
|
25
|
+
Sample G1 G2 G3 ... Gk
|
|
26
|
+
S1 0 1 0 0
|
|
27
|
+
S2 2 1 0 NA # exclude this sample
|
|
28
|
+
...
|
|
29
|
+
Sm 1 0 0 0
|
|
30
|
+
```
|
|
31
|
+
fmlfile: The formula file. The first column is grouping and the
|
|
32
|
+
second column is the formula. It must be tab-delimited.
|
|
33
|
+
```
|
|
34
|
+
Group Formula ... # Other columns to be added to outfile
|
|
35
|
+
G1 Fn ~ F1 + Fx + Fy # Fx, Fy could be covariates
|
|
36
|
+
G1 Fn ~ F2 + Fx + Fy
|
|
37
|
+
...
|
|
38
|
+
Gk Fn ~ F3 + Fx + Fy
|
|
39
|
+
```
|
|
40
|
+
|
|
41
|
+
Output:
|
|
42
|
+
outfile: The output file. It is a tab-delimited file with the first
|
|
43
|
+
column as the grouping and the second column as the p-value.
|
|
44
|
+
```
|
|
45
|
+
Group Formula ... Pooled Groups SSR SumSSR Fstat Pval Padj
|
|
46
|
+
G1 Fn ~ F1 0.123 2 1 0.123 0.123 0.123 0.123
|
|
47
|
+
G1 Fn ~ F2 0.123 2 1 0.123 0.123 0.123 0.123
|
|
48
|
+
...
|
|
49
|
+
Gk Fn ~ F3 0.123 2 1 0.123 0.123 0.123 0.123
|
|
50
|
+
```
|
|
51
|
+
|
|
52
|
+
Envs:
|
|
53
|
+
padj (choice): The method for p-value adjustment.
|
|
54
|
+
- none: No p-value adjustment (no Padj column in outfile).
|
|
55
|
+
- holm: Holm-Bonferroni method.
|
|
56
|
+
- hochberg: Hochberg method.
|
|
57
|
+
- hommel: Hommel method.
|
|
58
|
+
- bonferroni: Bonferroni method.
|
|
59
|
+
- BH: Benjamini-Hochberg method.
|
|
60
|
+
- BY: Benjamini-Yekutieli method.
|
|
61
|
+
- fdr: FDR correction method.
|
|
62
|
+
transpose_input (flag): Whether to transpose the input file.
|
|
63
|
+
transpose_group (flag): Whether to transpose the group file.
|
|
64
|
+
"""
|
|
65
|
+
input = "infile:file, groupfile:file, fmlfile:file"
|
|
66
|
+
output = "outfile:file:{{in.infile | stem}}.chowtest.txt"
|
|
67
|
+
lang = config.lang.rscript
|
|
68
|
+
envs = {
|
|
69
|
+
"padj": "none",
|
|
70
|
+
"transpose_input": False,
|
|
71
|
+
"transpose_group": False,
|
|
72
|
+
}
|
|
73
|
+
script = "file://../scripts/stats/ChowTest.R"
|
|
74
|
+
|
|
75
|
+
|
|
76
|
+
class Mediation(Proc):
|
|
77
|
+
"""Mediation analysis.
|
|
78
|
+
|
|
79
|
+
The flowchart of mediation analysis:
|
|
80
|
+
|
|
81
|
+
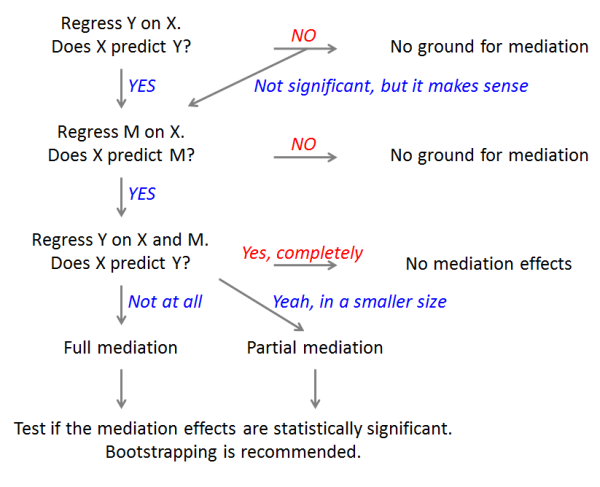
|
|
82
|
+
|
|
83
|
+
Reference:
|
|
84
|
+
- <https://library.virginia.edu/data/articles/introduction-to-mediation-analysis>
|
|
85
|
+
- <https://en.wikipedia.org/wiki/Mediation_(statistics)>
|
|
86
|
+
- <https://tilburgsciencehub.com/topics/analyze/regression/linear-regression/mediation-analysis/>
|
|
87
|
+
- <https://ademos.people.uic.edu/Chapter14.html>
|
|
88
|
+
|
|
89
|
+
Input:
|
|
90
|
+
infile: The input data file. The rows are samples and the columns are
|
|
91
|
+
features. It must be tab-delimited.
|
|
92
|
+
```
|
|
93
|
+
Sample F1 F2 F3 ... Fn
|
|
94
|
+
S1 1.2 3.4 5.6 7.8
|
|
95
|
+
S2 2.3 4.5 6.7 8.9
|
|
96
|
+
...
|
|
97
|
+
Sm 5.6 7.8 9.0 1.2
|
|
98
|
+
```
|
|
99
|
+
fmlfile: The formula file.
|
|
100
|
+
```
|
|
101
|
+
Case M Y X Cov Model_M Model_Y
|
|
102
|
+
Case1 F1 F2 F3 F4,F5 glm lm
|
|
103
|
+
...
|
|
104
|
+
```
|
|
105
|
+
Where Y is the outcome variable, X is the predictor variable, M is the
|
|
106
|
+
mediator variable, and Case is the case name. Model_M and Model_Y are the
|
|
107
|
+
models for M and Y, respectively.
|
|
108
|
+
`envs.cases` will be ignored if this is provided.
|
|
109
|
+
|
|
110
|
+
Output:
|
|
111
|
+
outfile: The output file.
|
|
112
|
+
Columns to help understand the results:
|
|
113
|
+
Total Effect: a total effect of X on Y (without M) (`Y ~ X`).
|
|
114
|
+
ADE: A Direct Effect of X on Y after taking into account a mediation effect of M (`Y ~ X + M`).
|
|
115
|
+
ACME: The Mediation Effect, the total effect minus the direct effect,
|
|
116
|
+
which equals to a product of a coefficient of X in the second step and a coefficient of M in the last step.
|
|
117
|
+
The goal of mediation analysis is to obtain this indirect effect and see if it's statistically significant.
|
|
118
|
+
|
|
119
|
+
Envs:
|
|
120
|
+
ncores (type=int): Number of cores to use for parallelization for cases.
|
|
121
|
+
sims (type=int): Number of Monte Carlo draws for nonparametric bootstrap or quasi-Bayesian approximation.
|
|
122
|
+
Will be passed to `mediation::mediate` function.
|
|
123
|
+
args (ns): Other arguments passed to `mediation::mediate` function.
|
|
124
|
+
- <more>: More arguments passed to `mediation::mediate` function.
|
|
125
|
+
See: <https://rdrr.io/cran/mediation/man/mediate.html>
|
|
126
|
+
padj (choice): The method for (ACME) p-value adjustment.
|
|
127
|
+
- none: No p-value adjustment (no Padj column in outfile).
|
|
128
|
+
- holm: Holm-Bonferroni method.
|
|
129
|
+
- hochberg: Hochberg method.
|
|
130
|
+
- hommel: Hommel method.
|
|
131
|
+
- bonferroni: Bonferroni method.
|
|
132
|
+
- BH: Benjamini-Hochberg method.
|
|
133
|
+
- BY: Benjamini-Yekutieli method.
|
|
134
|
+
- fdr: FDR correction method.
|
|
135
|
+
cases (type=json): The cases for mediation analysis.
|
|
136
|
+
Ignored if `in.fmlfile` is provided.
|
|
137
|
+
A json/dict with case names as keys and values as a dict of M, Y, X, Cov, Model_M, Model_Y.
|
|
138
|
+
For example:
|
|
139
|
+
```json
|
|
140
|
+
{
|
|
141
|
+
"Case1": {
|
|
142
|
+
"M": "F1",
|
|
143
|
+
"Y": "F2",
|
|
144
|
+
"X": "F3",
|
|
145
|
+
"Cov": "F4,F5",
|
|
146
|
+
"Model_M": "glm",
|
|
147
|
+
"Model_Y": "lm"
|
|
148
|
+
},
|
|
149
|
+
...
|
|
150
|
+
}
|
|
151
|
+
```
|
|
152
|
+
transpose_input (flag): Whether to transpose the input file.
|
|
153
|
+
""" # noqa: E501
|
|
154
|
+
input = "infile:file, fmlfile:file"
|
|
155
|
+
output = "outfile:file:{{in.infile | stem}}.mediation.txt"
|
|
156
|
+
lang = config.lang.rscript
|
|
157
|
+
envs = {
|
|
158
|
+
"ncores": config.misc.ncores,
|
|
159
|
+
"sims": 1000,
|
|
160
|
+
"args": {},
|
|
161
|
+
"padj": "none",
|
|
162
|
+
"cases": {},
|
|
163
|
+
"transpose_input": False,
|
|
164
|
+
}
|
|
165
|
+
script = "file://../scripts/stats/Mediation.R"
|
|
166
|
+
|
|
167
|
+
|
|
168
|
+
class LiquidAssoc(Proc):
|
|
169
|
+
"""Liquid association tests.
|
|
170
|
+
|
|
171
|
+
See Also https://github.com/gundt/fastLiquidAssociation
|
|
172
|
+
Requires https://github.com/pwwang/fastLiquidAssociation
|
|
173
|
+
|
|
174
|
+
Input:
|
|
175
|
+
infile: The input data file. The rows are samples and the columns are
|
|
176
|
+
features. It must be tab-delimited.
|
|
177
|
+
```
|
|
178
|
+
Sample F1 F2 F3 ... Fn
|
|
179
|
+
S1 1.2 3.4 5.6 7.8
|
|
180
|
+
S2 2.3 4.5 6.7 8.9
|
|
181
|
+
...
|
|
182
|
+
Sm 5.6 7.8 9.0 1.2
|
|
183
|
+
```
|
|
184
|
+
The features (columns) will be tested pairwise, which will be the X and
|
|
185
|
+
Y columns in the result of `fastMLA`
|
|
186
|
+
covfile: The covariate file. The rows are the samples and the columns
|
|
187
|
+
are the covariates. It must be tab-delimited.
|
|
188
|
+
If provided, the data in `in.infile` will be adjusted by covariates by
|
|
189
|
+
regressing out the covariates and the residuals will be used for
|
|
190
|
+
liquid association tests.
|
|
191
|
+
groupfile: The group file. The rows are the samples and the columns
|
|
192
|
+
are the groupings. It must be tab-delimited.
|
|
193
|
+
```
|
|
194
|
+
Sample G1 G2 G3 ... Gk
|
|
195
|
+
S1 0 1 0 0
|
|
196
|
+
S2 2 1 0 NA # exclude this sample
|
|
197
|
+
...
|
|
198
|
+
Sm 1 0 0 0
|
|
199
|
+
```
|
|
200
|
+
This will be served as the Z column in the result of `fastMLA`
|
|
201
|
+
This can be omitted. If so, `envs.nvec` should be specified, which is
|
|
202
|
+
to select column from `in.infile` as Z.
|
|
203
|
+
fmlfile: The formula file. The 3 columns are X3, X12 and X21. The results
|
|
204
|
+
will be filtered based on the formula. It must be tab-delimited without
|
|
205
|
+
header.
|
|
206
|
+
|
|
207
|
+
Output:
|
|
208
|
+
outfile: The output file.
|
|
209
|
+
```
|
|
210
|
+
X12 X21 X3 rhodiff MLA value estimates san.se wald Pval model
|
|
211
|
+
C38 C46 C5 0.87 0.32 0.67 0.20 10.87 0 F
|
|
212
|
+
C46 C38 C5 0.87 0.32 0.67 0.20 10.87 0 F
|
|
213
|
+
C27 C39 C4 0.94 0.34 1.22 0.38 10.03 0 F
|
|
214
|
+
```
|
|
215
|
+
|
|
216
|
+
Envs:
|
|
217
|
+
nvec: The column index (1-based) of Z in `in.infile`, if `in.groupfile` is
|
|
218
|
+
omitted. You can specify multiple columns by comma-seperated values, or
|
|
219
|
+
a range of columns by `-`. For example, `1,3,5-7,9`. It also supports
|
|
220
|
+
column names. For example, `F1,F3`. `-` is not supported for column
|
|
221
|
+
names.
|
|
222
|
+
x: Similar as `nvec`, but limit X group to given features.
|
|
223
|
+
The rest of features (other than X and Z) in `in.infile` will
|
|
224
|
+
be used as Y.
|
|
225
|
+
The features in `in.infile` will still be tested pairwise, but only
|
|
226
|
+
features in X and Y will be kept.
|
|
227
|
+
topn (type=int): Number of results to return by `fastMLA`, ordered from
|
|
228
|
+
highest `|MLA|` value descending.
|
|
229
|
+
The default of the package is 2000, but here we set to 1e6 to return as
|
|
230
|
+
many results as possible (also good to do pvalue adjustment).
|
|
231
|
+
rvalue (type=float): Tolerance value for LA approximation. Lower values of
|
|
232
|
+
rvalue will cause a more thorough search, but take longer.
|
|
233
|
+
cut (type=int): Value passed to the GLA function to create buckets
|
|
234
|
+
(equal to number of buckets+1). Values placing between 15-30 samples per
|
|
235
|
+
bucket are optimal. Must be a positive integer>1. By default,
|
|
236
|
+
`max(ceiling(nrow(data)/22), 4)` is used.
|
|
237
|
+
ncores (type=int): Number of cores to use for parallelization.
|
|
238
|
+
padj (choice): The method for p-value adjustment.
|
|
239
|
+
- none: No p-value adjustment (no Padj column in outfile).
|
|
240
|
+
- holm: Holm-Bonferroni method.
|
|
241
|
+
- hochberg: Hochberg method.
|
|
242
|
+
- hommel: Hommel method.
|
|
243
|
+
- bonferroni: Bonferroni method.
|
|
244
|
+
- BH: Benjamini-Hochberg method.
|
|
245
|
+
- BY: Benjamini-Yekutieli method.
|
|
246
|
+
- fdr: FDR correction method.
|
|
247
|
+
transpose_input (flag): Whether to transpose the input file.
|
|
248
|
+
transpose_group (flag): Whether to transpose the group file.
|
|
249
|
+
transpose_cov (flag): Whether to transpose the covariate file.
|
|
250
|
+
xyz_names: The names of X12, X21 and X3 in the final output file. Separated
|
|
251
|
+
by comma. For example, `X12,X21,X3`.
|
|
252
|
+
"""
|
|
253
|
+
input = "infile:file, covfile:file, groupfile:file, fmlfile:file"
|
|
254
|
+
output = "outfile:file:{{in.infile | stem}}.liquidassoc.txt"
|
|
255
|
+
lang = config.lang.rscript
|
|
256
|
+
envs = {
|
|
257
|
+
"nvec": None,
|
|
258
|
+
"x": None,
|
|
259
|
+
"topn": 1e6,
|
|
260
|
+
"rvalue": 0.5,
|
|
261
|
+
"cut": 20,
|
|
262
|
+
"ncores": config.misc.ncores,
|
|
263
|
+
"padj": "none",
|
|
264
|
+
"transpose_input": False,
|
|
265
|
+
"transpose_group": False,
|
|
266
|
+
"transpose_cov": False,
|
|
267
|
+
"xyz_names": None,
|
|
268
|
+
}
|
|
269
|
+
script = "file://../scripts/stats/LiquidAssoc.R"
|
|
270
|
+
|
|
271
|
+
|
|
272
|
+
class DiffCoexpr(Proc):
|
|
273
|
+
"""Differential co-expression analysis.
|
|
274
|
+
|
|
275
|
+
See also <https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-11-497>
|
|
276
|
+
and <https://github.com/DavisLaboratory/dcanr/blob/8958d61788937eef3b7e2b4118651cbd7af7469d/R/inference_methods.R#L199>.
|
|
277
|
+
|
|
278
|
+
Input:
|
|
279
|
+
infile: The input data file. The rows are samples and the columns are
|
|
280
|
+
features. It must be tab-delimited.
|
|
281
|
+
```
|
|
282
|
+
Sample F1 F2 F3 ... Fn
|
|
283
|
+
S1 1.2 3.4 5.6 7.8
|
|
284
|
+
S2 2.3 4.5 6.7 8.9
|
|
285
|
+
...
|
|
286
|
+
Sm 5.6 7.8 9.0 1.2
|
|
287
|
+
```
|
|
288
|
+
groupfile: The group file. The rows are the samples and the columns
|
|
289
|
+
are the groupings. It must be tab-delimited.
|
|
290
|
+
```
|
|
291
|
+
Sample G1 G2 G3 ... Gk
|
|
292
|
+
S1 0 1 0 0
|
|
293
|
+
S2 2 1 0 NA # exclude this sample
|
|
294
|
+
...
|
|
295
|
+
Sm 1 0 0 0
|
|
296
|
+
```
|
|
297
|
+
|
|
298
|
+
Output:
|
|
299
|
+
outfile: The output file. It is a tab-delimited file with the first
|
|
300
|
+
column as the feature pair and the second column as the p-value.
|
|
301
|
+
```
|
|
302
|
+
Group Feature1 Feature2 Pval Padj
|
|
303
|
+
G1 F1 F2 0.123 0.123
|
|
304
|
+
G1 F1 F3 0.123 0.123
|
|
305
|
+
...
|
|
306
|
+
```
|
|
307
|
+
|
|
308
|
+
Envs:
|
|
309
|
+
method (choice): The method used to calculate the differential
|
|
310
|
+
co-expression.
|
|
311
|
+
- pearson: Pearson correlation.
|
|
312
|
+
- spearman: Spearman correlation.
|
|
313
|
+
beta: The beta value for the differential co-expression analysis.
|
|
314
|
+
padj (choice): The method for p-value adjustment.
|
|
315
|
+
- none: No p-value adjustment (no Padj column in outfile).
|
|
316
|
+
- holm: Holm-Bonferroni method.
|
|
317
|
+
- hochberg: Hochberg method.
|
|
318
|
+
- hommel: Hommel method.
|
|
319
|
+
- bonferroni: Bonferroni method.
|
|
320
|
+
- BH: Benjamini-Hochberg method.
|
|
321
|
+
- BY: Benjamini-Yekutieli method.
|
|
322
|
+
- fdr: FDR correction method.
|
|
323
|
+
perm_batch (type=int): The number of permutations to run in each batch
|
|
324
|
+
seed (type=int): The seed for random number generation
|
|
325
|
+
ncores (type=int): The number of cores to use for parallelization
|
|
326
|
+
transpose_input (flag): Whether to transpose the input file.
|
|
327
|
+
transpose_group (flag): Whether to transpose the group file.
|
|
328
|
+
""" # noqa: E501
|
|
329
|
+
input = "infile:file, groupfile:file"
|
|
330
|
+
output = "outfile:file:{{in.infile | stem}}.diffcoexpr.txt"
|
|
331
|
+
lang = config.lang.rscript
|
|
332
|
+
envs = {
|
|
333
|
+
"method": "pearson",
|
|
334
|
+
"beta": 6,
|
|
335
|
+
"padj": "none",
|
|
336
|
+
"perm_batch": 20,
|
|
337
|
+
"seed": 8525,
|
|
338
|
+
"ncores": config.misc.ncores,
|
|
339
|
+
"transpose_input": False,
|
|
340
|
+
"transpose_group": False,
|
|
341
|
+
}
|
|
342
|
+
script = "file://../scripts/stats/DiffCoexpr.R"
|
|
343
|
+
|
|
344
|
+
|
|
345
|
+
class MetaPvalue(Proc):
|
|
346
|
+
"""Calulation of meta p-values.
|
|
347
|
+
|
|
348
|
+
If there is only one input file, only the p-value adjustment will be performed.
|
|
349
|
+
|
|
350
|
+
Input:
|
|
351
|
+
infiles: The input files. Each file is a tab-delimited file with multiple
|
|
352
|
+
columns. There should be ID column(s) to match the rows in other files and
|
|
353
|
+
p-value column(s) to be combined. The records will be full-joined by ID.
|
|
354
|
+
When only one file is provided, only the pvalue adjustment will be
|
|
355
|
+
performed when `envs.padj` is not `none`, otherwise the input file will
|
|
356
|
+
be copied to `out.outfile`.
|
|
357
|
+
|
|
358
|
+
Output:
|
|
359
|
+
outfile: The output file. It is a tab-delimited file with the first column as
|
|
360
|
+
the ID and the second column as the combined p-value.
|
|
361
|
+
```
|
|
362
|
+
ID ID1 ... Pval Padj
|
|
363
|
+
a x ... 0.123 0.123
|
|
364
|
+
b y ... 0.123 0.123
|
|
365
|
+
...
|
|
366
|
+
```
|
|
367
|
+
|
|
368
|
+
Envs:
|
|
369
|
+
id_cols: The column names used in all `in.infiles` as ID columns. Multiple
|
|
370
|
+
columns can be specified by comma-seperated values. For example, `ID1,ID2`,
|
|
371
|
+
where `ID1` is the ID column in the first file and `ID2` is the ID column
|
|
372
|
+
in the second file.
|
|
373
|
+
If `id_exprs` is specified, this should be a single column name for the new
|
|
374
|
+
ID column in each `in.infiles` and the final `out.outfile`.
|
|
375
|
+
id_exprs: The R expressions for each `in.infiles` to get ID column(s).
|
|
376
|
+
pval_cols: The column names used in all `in.infiles` as p-value columns.
|
|
377
|
+
Different columns can be specified by comma-seperated values for each
|
|
378
|
+
`in.infiles`. For example, `Pval1,Pval2`.
|
|
379
|
+
method (choice): The method used to calculate the meta-pvalue.
|
|
380
|
+
- fisher: Fisher's method.
|
|
381
|
+
- sumlog: Sum of logarithms (same as Fisher's method)
|
|
382
|
+
- logitp: Logit method.
|
|
383
|
+
- sumz: Sum of z method (Stouffer's method).
|
|
384
|
+
- meanz: Mean of z method.
|
|
385
|
+
- meanp: Mean of p method.
|
|
386
|
+
- invt: Inverse t method.
|
|
387
|
+
- sump: Sum of p method (Edgington's method).
|
|
388
|
+
- votep: Vote counting method.
|
|
389
|
+
- wilkinsonp: Wilkinson's method.
|
|
390
|
+
- invchisq: Inverse chi-square method.
|
|
391
|
+
keep_single (flag): Whether to keep the original p-value when there is only one
|
|
392
|
+
p-value.
|
|
393
|
+
na: The method to handle NA values. -1 to skip the record. Otherwise NA
|
|
394
|
+
will be replaced by the given value.
|
|
395
|
+
padj (choice): The method for p-value adjustment.
|
|
396
|
+
- none: No p-value adjustment (no Padj column in outfile).
|
|
397
|
+
- holm: Holm-Bonferroni method.
|
|
398
|
+
- hochberg: Hochberg method.
|
|
399
|
+
- hommel: Hommel method.
|
|
400
|
+
- bonferroni: Bonferroni method.
|
|
401
|
+
- BH: Benjamini-Hochberg method.
|
|
402
|
+
- BY: Benjamini-Yekutieli method.
|
|
403
|
+
- fdr: FDR correction method.
|
|
404
|
+
"""
|
|
405
|
+
input = "infiles:files"
|
|
406
|
+
output = "outfile:file:{{in.infiles | first | stem}}.metapval.txt"
|
|
407
|
+
lang = config.lang.rscript
|
|
408
|
+
envs = {
|
|
409
|
+
"id_cols": None,
|
|
410
|
+
"id_exprs": None,
|
|
411
|
+
"pval_cols": None,
|
|
412
|
+
"method": "fisher",
|
|
413
|
+
"na": -1,
|
|
414
|
+
"keep_single": True,
|
|
415
|
+
"padj": "none",
|
|
416
|
+
}
|
|
417
|
+
script = "file://../scripts/stats/MetaPvalue.R"
|
|
418
|
+
|
|
419
|
+
|
|
420
|
+
class MetaPvalue1(Proc):
|
|
421
|
+
"""Calulation of meta p-values.
|
|
422
|
+
|
|
423
|
+
Unlike `MetaPvalue`, this process only accepts one input file.
|
|
424
|
+
|
|
425
|
+
The p-values will be grouped by the ID columns and combined by the selected method.
|
|
426
|
+
|
|
427
|
+
Input:
|
|
428
|
+
infile: The input file.
|
|
429
|
+
The file is a tab-delimited file with multiple
|
|
430
|
+
columns. There should be ID column(s) to group the rows where
|
|
431
|
+
p-value column(s) to be combined.
|
|
432
|
+
|
|
433
|
+
Output:
|
|
434
|
+
outfile: The output file. It is a tab-delimited file with the first column as
|
|
435
|
+
the ID and the second column as the combined p-value.
|
|
436
|
+
```
|
|
437
|
+
ID ID1 ... Pval Padj
|
|
438
|
+
a x ... 0.123 0.123
|
|
439
|
+
b y ... 0.123 0.123
|
|
440
|
+
...
|
|
441
|
+
```
|
|
442
|
+
|
|
443
|
+
Envs:
|
|
444
|
+
id_cols: The column names used in `in.infile` as ID columns. Multiple
|
|
445
|
+
columns can be specified by comma-seperated values. For example, `ID1,ID2`.
|
|
446
|
+
pval_col: The column name used in `in.infile` as p-value column.
|
|
447
|
+
method (choice): The method used to calculate the meta-pvalue.
|
|
448
|
+
- fisher: Fisher's method.
|
|
449
|
+
- sumlog: Sum of logarithms (same as Fisher's method)
|
|
450
|
+
- logitp: Logit method.
|
|
451
|
+
- sumz: Sum of z method (Stouffer's method).
|
|
452
|
+
- meanz: Mean of z method.
|
|
453
|
+
- meanp: Mean of p method.
|
|
454
|
+
- invt: Inverse t method.
|
|
455
|
+
- sump: Sum of p method (Edgington's method).
|
|
456
|
+
- votep: Vote counting method.
|
|
457
|
+
- wilkinsonp: Wilkinson's method.
|
|
458
|
+
- invchisq: Inverse chi-square method.
|
|
459
|
+
na: The method to handle NA values. -1 to skip the record. Otherwise NA
|
|
460
|
+
will be replaced by the given value.
|
|
461
|
+
keep_single (flag): Whether to keep the original p-value when there is only one
|
|
462
|
+
p-value.
|
|
463
|
+
padj (choice): The method for p-value adjustment.
|
|
464
|
+
- none: No p-value adjustment (no Padj column in outfile).
|
|
465
|
+
- holm: Holm-Bonferroni method.
|
|
466
|
+
- hochberg: Hochberg method.
|
|
467
|
+
- hommel: Hommel method.
|
|
468
|
+
- bonferroni: Bonferroni method.
|
|
469
|
+
- BH: Benjamini-Hochberg method.
|
|
470
|
+
- BY: Benjamini-Yekutieli method.
|
|
471
|
+
- fdr: FDR correction method.
|
|
472
|
+
"""
|
|
473
|
+
input = "infile:file"
|
|
474
|
+
output = "outfile:file:{{in.infile | stem}}.metapval.txt"
|
|
475
|
+
lang = config.lang.rscript
|
|
476
|
+
envs = {
|
|
477
|
+
"id_cols": None,
|
|
478
|
+
"pval_col": None,
|
|
479
|
+
"method": "fisher",
|
|
480
|
+
"na": -1,
|
|
481
|
+
"keep_single": True,
|
|
482
|
+
"padj": "none",
|
|
483
|
+
}
|
|
484
|
+
script = "file://../scripts/stats/MetaPvalue1.R"
|