microlive 1.0.11__py3-none-any.whl
This diff represents the content of publicly available package versions that have been released to one of the supported registries. The information contained in this diff is provided for informational purposes only and reflects changes between package versions as they appear in their respective public registries.
- microlive/__init__.py +50 -0
- microlive/data/__init__.py +0 -0
- microlive/data/icons/__init__.py +0 -0
- microlive/data/icons/icon_micro.png +0 -0
- microlive/data/models/__init__.py +0 -0
- microlive/gui/__init__.py +1 -0
- microlive/gui/app.py +16340 -0
- microlive/gui/main.py +86 -0
- microlive/gui/micro_mac.command +18 -0
- microlive/gui/micro_windows.bat +24 -0
- microlive/imports.py +209 -0
- microlive/microscopy.py +13321 -0
- microlive/ml_spot_detection.py +252 -0
- microlive/pipelines/__init__.py +17 -0
- microlive/pipelines/pipeline_FRAP.py +1225 -0
- microlive/pipelines/pipeline_folding_efficiency.py +297 -0
- microlive/pipelines/pipeline_particle_tracking.py +489 -0
- microlive/pipelines/pipeline_spot_detection_no_tracking.py +368 -0
- microlive/utils/__init__.py +13 -0
- microlive/utils/device.py +99 -0
- microlive/utils/resources.py +60 -0
- microlive-1.0.11.dist-info/METADATA +361 -0
- microlive-1.0.11.dist-info/RECORD +26 -0
- microlive-1.0.11.dist-info/WHEEL +4 -0
- microlive-1.0.11.dist-info/entry_points.txt +2 -0
- microlive-1.0.11.dist-info/licenses/LICENSE +674 -0
|
@@ -0,0 +1,361 @@
|
|
|
1
|
+
Metadata-Version: 2.4
|
|
2
|
+
Name: microlive
|
|
3
|
+
Version: 1.0.11
|
|
4
|
+
Summary: Live-cell microscopy image analysis and single-molecule measurements
|
|
5
|
+
Project-URL: Homepage, https://github.com/ningzhaoAnschutz/microlive
|
|
6
|
+
Project-URL: Documentation, https://github.com/ningzhaoAnschutz/microlive/blob/main/docs/user_guide.md
|
|
7
|
+
Project-URL: Repository, https://github.com/ningzhaoAnschutz/microlive
|
|
8
|
+
Project-URL: Issues, https://github.com/ningzhaoAnschutz/microlive/issues
|
|
9
|
+
Project-URL: Changelog, https://github.com/ningzhaoAnschutz/microlive/releases
|
|
10
|
+
Author: Luis U. Aguilera, William S. Raymond, Rhiannon M. Sears, Nathan L. Nowling, Brian Munsky, Ning Zhao
|
|
11
|
+
License-Expression: GPL-3.0-or-later
|
|
12
|
+
License-File: LICENSE
|
|
13
|
+
Keywords: PyQt5,bioimaging,cellpose,image-analysis,live-cell,microscopy,particle-tracking,single-molecule,tracking
|
|
14
|
+
Classifier: Development Status :: 4 - Beta
|
|
15
|
+
Classifier: Environment :: X11 Applications :: Qt
|
|
16
|
+
Classifier: Intended Audience :: Science/Research
|
|
17
|
+
Classifier: License :: OSI Approved :: GNU General Public License v3 (GPLv3)
|
|
18
|
+
Classifier: Operating System :: MacOS :: MacOS X
|
|
19
|
+
Classifier: Operating System :: Microsoft :: Windows
|
|
20
|
+
Classifier: Operating System :: OS Independent
|
|
21
|
+
Classifier: Operating System :: POSIX :: Linux
|
|
22
|
+
Classifier: Programming Language :: Python :: 3
|
|
23
|
+
Classifier: Programming Language :: Python :: 3.10
|
|
24
|
+
Classifier: Programming Language :: Python :: 3.11
|
|
25
|
+
Classifier: Programming Language :: Python :: 3.12
|
|
26
|
+
Classifier: Topic :: Scientific/Engineering :: Bio-Informatics
|
|
27
|
+
Classifier: Topic :: Scientific/Engineering :: Image Processing
|
|
28
|
+
Classifier: Topic :: Scientific/Engineering :: Visualization
|
|
29
|
+
Requires-Python: >=3.10
|
|
30
|
+
Requires-Dist: big-fish>=0.6
|
|
31
|
+
Requires-Dist: bioio-base==3.1.0
|
|
32
|
+
Requires-Dist: bioio-lif>=1.2
|
|
33
|
+
Requires-Dist: bioio==3.1.0
|
|
34
|
+
Requires-Dist: biopython>=1.81
|
|
35
|
+
Requires-Dist: brokenaxes
|
|
36
|
+
Requires-Dist: cellpose>=3.0
|
|
37
|
+
Requires-Dist: dask==2024.8.0
|
|
38
|
+
Requires-Dist: distributed==2024.8.0
|
|
39
|
+
Requires-Dist: dna-features-viewer>=3.1
|
|
40
|
+
Requires-Dist: fpdf>=1.7
|
|
41
|
+
Requires-Dist: imagecodecs
|
|
42
|
+
Requires-Dist: imageio>=2.34
|
|
43
|
+
Requires-Dist: ipython
|
|
44
|
+
Requires-Dist: ipywidgets
|
|
45
|
+
Requires-Dist: joblib>=1.3
|
|
46
|
+
Requires-Dist: joypy
|
|
47
|
+
Requires-Dist: llvmlite==0.43.0
|
|
48
|
+
Requires-Dist: matplotlib-scalebar
|
|
49
|
+
Requires-Dist: matplotlib==3.9.2
|
|
50
|
+
Requires-Dist: natsort
|
|
51
|
+
Requires-Dist: numba==0.60.0
|
|
52
|
+
Requires-Dist: numpy==1.26.4
|
|
53
|
+
Requires-Dist: opencv-python-headless>=4.9
|
|
54
|
+
Requires-Dist: openpyxl>=3.1
|
|
55
|
+
Requires-Dist: pandas==2.2.2
|
|
56
|
+
Requires-Dist: pillow>=10.0
|
|
57
|
+
Requires-Dist: psutil
|
|
58
|
+
Requires-Dist: pydantic>=2.0
|
|
59
|
+
Requires-Dist: pyqt5>=5.15
|
|
60
|
+
Requires-Dist: pystackreg>=0.2.8
|
|
61
|
+
Requires-Dist: pyyaml>=6.0
|
|
62
|
+
Requires-Dist: readlif==0.6.5
|
|
63
|
+
Requires-Dist: roifile
|
|
64
|
+
Requires-Dist: scikit-image==0.24.0
|
|
65
|
+
Requires-Dist: scikit-learn==1.5.2
|
|
66
|
+
Requires-Dist: scipy==1.13.1
|
|
67
|
+
Requires-Dist: seaborn==0.13.2
|
|
68
|
+
Requires-Dist: snapgene-reader>=0.1
|
|
69
|
+
Requires-Dist: statsmodels==0.14.4
|
|
70
|
+
Requires-Dist: tifffile>=2024.1
|
|
71
|
+
Requires-Dist: tqdm>=4.66
|
|
72
|
+
Requires-Dist: trackpy>=0.6
|
|
73
|
+
Provides-Extra: dev
|
|
74
|
+
Requires-Dist: black>=23.0; extra == 'dev'
|
|
75
|
+
Requires-Dist: build; extra == 'dev'
|
|
76
|
+
Requires-Dist: isort>=5.12; extra == 'dev'
|
|
77
|
+
Requires-Dist: mypy>=1.5; extra == 'dev'
|
|
78
|
+
Requires-Dist: pdoc; extra == 'dev'
|
|
79
|
+
Requires-Dist: pytest-qt>=4.0; extra == 'dev'
|
|
80
|
+
Requires-Dist: pytest>=7.0; extra == 'dev'
|
|
81
|
+
Requires-Dist: ruff>=0.1; extra == 'dev'
|
|
82
|
+
Requires-Dist: twine; extra == 'dev'
|
|
83
|
+
Provides-Extra: notebooks
|
|
84
|
+
Requires-Dist: bqplot; extra == 'notebooks'
|
|
85
|
+
Requires-Dist: ipympl>=0.9; extra == 'notebooks'
|
|
86
|
+
Requires-Dist: ipywidgets>=8.0; extra == 'notebooks'
|
|
87
|
+
Requires-Dist: jupyter>=1.0; extra == 'notebooks'
|
|
88
|
+
Description-Content-Type: text/markdown
|
|
89
|
+
|
|
90
|
+
# MicroLive <img src="https://raw.githubusercontent.com/ningzhaoAnschutz/microlive/main/docs/icons/icon_micro.png" alt="Micro Logo" width="150" align="right" />
|
|
91
|
+
|
|
92
|
+
**Authors:** Luis U. Aguilera, William S. Raymond, Rhiannon M. Sears, Nathan L. Nowling, Brian Munsky, Ning Zhao
|
|
93
|
+
|
|
94
|
+
[](https://pypi.org/project/microlive/) [](docs/user_guide.md) [](docs/tutorial.md) [](docs/api_reference.md) [](https://www.gnu.org/licenses/gpl-3.0) [](https://www.python.org/) [](https://pypi.org/project/PyQt5/)
|
|
95
|
+
|
|
96
|
+
## About
|
|
97
|
+
|
|
98
|
+
**MicroLive** is a Python-based GUI application for live-cell microscopy image analysis and single-molecule measurements. It provides an end-to-end workflow from image loading through particle tracking, colocalization analysis, and statistical analysis.
|
|
99
|
+
|
|
100
|
+
[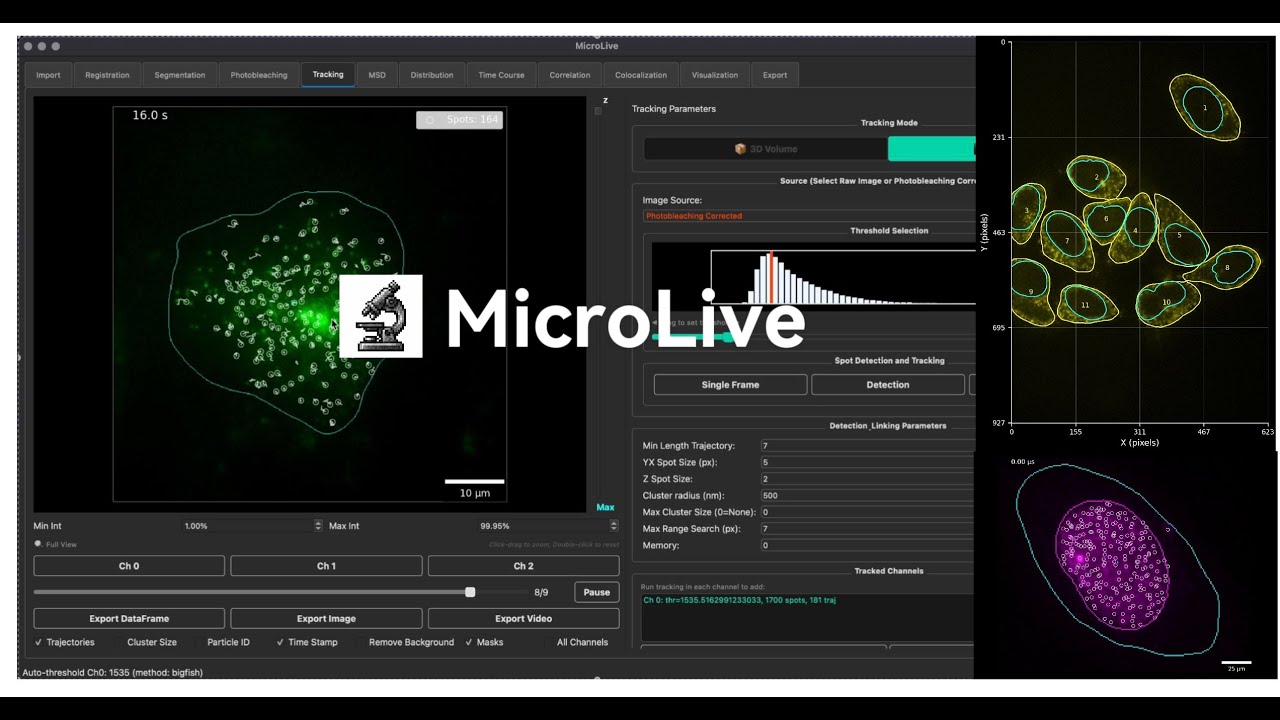](https://youtu.be/GU1ct5go-AU?si=wEJWUaSYgNGj-Pi3)
|
|
101
|
+
|
|
102
|
+
*Click the image above to watch the demo video on YouTube*
|
|
103
|
+
|
|
104
|
+
---
|
|
105
|
+
|
|
106
|
+
## Features
|
|
107
|
+
|
|
108
|
+
- **Image I/O**: Load .lif, .tif, .ome.tif with metadata extraction and dimension mapping
|
|
109
|
+
- **Registration**: Drift correction via phase correlation
|
|
110
|
+
- **Segmentation**: Cellpose (GPU), watershed, manual ROI, or external mask import
|
|
111
|
+
- **Photobleaching correction**: Exponential decay modeling
|
|
112
|
+
- **Particle tracking**: 2D (TrackPy) and 3D (Big-FISH) detection with multi-channel support
|
|
113
|
+
- **Automated threshold detection**: Hybrid Big-FISH/TrueSpot method
|
|
114
|
+
- **Trajectory linking**: Nearest-neighbor with memory and cluster analysis
|
|
115
|
+
- **Intensity quantification**: Background subtraction, PSF fitting, SNR calculation
|
|
116
|
+
- **Colocalization**: CNN-based, distance-based, and manual verification
|
|
117
|
+
- **MSD analysis**: Per-cell diffusion coefficient calculation
|
|
118
|
+
- **Correlation**: Auto- and cross-correlation with exponential/linear fitting
|
|
119
|
+
- **Export**: PNG, TIFF, CSV, MP4/GIF with full metadata logging
|
|
120
|
+
|
|
121
|
+
---
|
|
122
|
+
|
|
123
|
+
## Documentation
|
|
124
|
+
|
|
125
|
+
- **[User Guide](docs/user_guide.md)** — Complete guide to using MicroLive
|
|
126
|
+
- **[Tutorial](docs/tutorial.md)** — Step-by-step tutorials for all workflows
|
|
127
|
+
- **[API Reference](docs/api_reference.md)** — Technical documentation for developers
|
|
128
|
+
|
|
129
|
+
---
|
|
130
|
+
|
|
131
|
+
## Installation
|
|
132
|
+
|
|
133
|
+
### Quick Install (Recommended)
|
|
134
|
+
|
|
135
|
+
```bash
|
|
136
|
+
# Create and activate conda environment
|
|
137
|
+
conda create -n microlive python=3.10 -y
|
|
138
|
+
conda activate microlive
|
|
139
|
+
|
|
140
|
+
# Install MicroLive
|
|
141
|
+
pip install microlive
|
|
142
|
+
|
|
143
|
+
# Launch the GUI
|
|
144
|
+
microlive
|
|
145
|
+
```
|
|
146
|
+
|
|
147
|
+
That's it! MicroLive will launch with GPU acceleration automatically enabled:
|
|
148
|
+
|
|
149
|
+
- **macOS (Apple Silicon)**: MPS GPU acceleration works automatically
|
|
150
|
+
- **Linux/Windows (CPU)**: Works out of the box
|
|
151
|
+
|
|
152
|
+
---
|
|
153
|
+
|
|
154
|
+
### GPU Acceleration for NVIDIA (Windows/Linux)
|
|
155
|
+
|
|
156
|
+
For NVIDIA GPU acceleration, install PyTorch with CUDA support **before** installing MicroLive:
|
|
157
|
+
|
|
158
|
+
```bash
|
|
159
|
+
# Create environment
|
|
160
|
+
conda create -n microlive python=3.10 -y
|
|
161
|
+
conda activate microlive
|
|
162
|
+
|
|
163
|
+
# Install PyTorch with CUDA 12.4 (adjust version as needed)
|
|
164
|
+
pip install torch torchvision --index-url https://download.pytorch.org/whl/cu124
|
|
165
|
+
|
|
166
|
+
# Then install MicroLive
|
|
167
|
+
pip install microlive
|
|
168
|
+
|
|
169
|
+
# Launch
|
|
170
|
+
microlive
|
|
171
|
+
```
|
|
172
|
+
|
|
173
|
+
### Using Conda Environment Files
|
|
174
|
+
|
|
175
|
+
Pre-configured environment files are available:
|
|
176
|
+
|
|
177
|
+
```bash
|
|
178
|
+
# macOS / CPU
|
|
179
|
+
conda env create -f installation/microlive.yml
|
|
180
|
+
conda activate microlive
|
|
181
|
+
|
|
182
|
+
# Windows/Linux with NVIDIA GPU
|
|
183
|
+
conda env create -f installation/microlive_cuda.yml
|
|
184
|
+
conda activate microlive
|
|
185
|
+
```
|
|
186
|
+
|
|
187
|
+
### Verify GPU Support
|
|
188
|
+
|
|
189
|
+
```python
|
|
190
|
+
from microlive.utils.device import check_gpu_status
|
|
191
|
+
check_gpu_status()
|
|
192
|
+
```
|
|
193
|
+
|
|
194
|
+
Expected output:
|
|
195
|
+
|
|
196
|
+
```
|
|
197
|
+
PyTorch version: 2.x.x
|
|
198
|
+
✅ CUDA available: NVIDIA GeForce RTX ...
|
|
199
|
+
Memory: 8.0 GB
|
|
200
|
+
```
|
|
201
|
+
|
|
202
|
+
Or for Apple Silicon:
|
|
203
|
+
|
|
204
|
+
```
|
|
205
|
+
PyTorch version: 2.x.x
|
|
206
|
+
✅ MPS available: Apple Silicon GPU (MPS)
|
|
207
|
+
```
|
|
208
|
+
|
|
209
|
+
---
|
|
210
|
+
|
|
211
|
+
### Development Installation
|
|
212
|
+
|
|
213
|
+
For developers who want to modify the source code:
|
|
214
|
+
|
|
215
|
+
```bash
|
|
216
|
+
# Clone the repository
|
|
217
|
+
git clone --depth 1 https://github.com/ningzhaoAnschutz/microlive.git
|
|
218
|
+
cd microlive
|
|
219
|
+
|
|
220
|
+
# Create environment
|
|
221
|
+
conda create -n microlive python=3.10 -y
|
|
222
|
+
conda activate microlive
|
|
223
|
+
|
|
224
|
+
# Install in editable mode
|
|
225
|
+
pip install -e .
|
|
226
|
+
|
|
227
|
+
# Launch
|
|
228
|
+
microlive
|
|
229
|
+
```
|
|
230
|
+
|
|
231
|
+
---
|
|
232
|
+
|
|
233
|
+
### Troubleshooting Installation
|
|
234
|
+
|
|
235
|
+
If you encounter errors during installation (especially on Python 3.12):
|
|
236
|
+
|
|
237
|
+
```bash
|
|
238
|
+
# Option 1: Upgrade pip and pre-install numpy
|
|
239
|
+
pip install --upgrade pip setuptools wheel
|
|
240
|
+
pip install numpy
|
|
241
|
+
pip install microlive
|
|
242
|
+
|
|
243
|
+
# Option 2: Use Python 3.10 (most compatible)
|
|
244
|
+
conda create -n microlive python=3.10 -y
|
|
245
|
+
conda activate microlive
|
|
246
|
+
pip install microlive
|
|
247
|
+
```
|
|
248
|
+
|
|
249
|
+
**Common error**: `ModuleNotFoundError: No module named 'numpy'` during `pystackreg` build
|
|
250
|
+
|
|
251
|
+
- This occurs when pip tries to build older package versions from source
|
|
252
|
+
- Solution: Pre-install NumPy before installing MicroLive
|
|
253
|
+
|
|
254
|
+
---
|
|
255
|
+
|
|
256
|
+
## Usage
|
|
257
|
+
|
|
258
|
+
### GUI Application
|
|
259
|
+
|
|
260
|
+
```bash
|
|
261
|
+
conda activate microlive
|
|
262
|
+
microlive
|
|
263
|
+
```
|
|
264
|
+
|
|
265
|
+
### Programmatic API
|
|
266
|
+
|
|
267
|
+
```python
|
|
268
|
+
import microlive.microscopy as mi
|
|
269
|
+
|
|
270
|
+
# Load images from a Leica .lif file
|
|
271
|
+
reader = mi.ReadLif("experiment.lif")
|
|
272
|
+
images = reader.get_images()
|
|
273
|
+
|
|
274
|
+
# Access image data
|
|
275
|
+
image = reader.list_images[0]
|
|
276
|
+
print(f"Image shape: {image.shape}") # (T, Z, Y, X, C)
|
|
277
|
+
|
|
278
|
+
# Run Cellpose segmentation
|
|
279
|
+
cellpose = mi.Cellpose(image_for_segmentation=image[0, 0, :, :, 0])
|
|
280
|
+
masks = cellpose.calculate_masks()
|
|
281
|
+
|
|
282
|
+
# Spot detection
|
|
283
|
+
spots = mi.SpotDetection(
|
|
284
|
+
image=image,
|
|
285
|
+
spot_size_yx=5,
|
|
286
|
+
threshold=100
|
|
287
|
+
)
|
|
288
|
+
detected = spots.detect_spots()
|
|
289
|
+
```
|
|
290
|
+
|
|
291
|
+
---
|
|
292
|
+
|
|
293
|
+
## Project Structure
|
|
294
|
+
|
|
295
|
+
```text
|
|
296
|
+
microlive/
|
|
297
|
+
├── microlive/ # Core package (pip installable)
|
|
298
|
+
│ ├── microscopy.py # Main analysis classes
|
|
299
|
+
│ ├── imports.py # Central import management
|
|
300
|
+
│ ├── ml_spot_detection.py # ML-based spot detection
|
|
301
|
+
│ ├── gui/ # GUI application
|
|
302
|
+
│ │ ├── app.py # Main GUI window
|
|
303
|
+
│ │ └── main.py # CLI entry point
|
|
304
|
+
│ ├── utils/ # Utility modules
|
|
305
|
+
│ │ ├── device.py # GPU detection
|
|
306
|
+
│ │ └── resources.py # Resource paths
|
|
307
|
+
│ ├── data/ # ML models and resources
|
|
308
|
+
│ └── pipelines/ # Analysis pipeline modules
|
|
309
|
+
├── simulations/ # Spot simulation & validation (not in pip)
|
|
310
|
+
│ ├── spot_simulator.py # Multi-cell simulation engine
|
|
311
|
+
│ ├── run_simulation.py # CLI runner
|
|
312
|
+
│ ├── visualize_results.py # Visualization tools
|
|
313
|
+
│ ├── config_*.yaml # Configuration examples
|
|
314
|
+
│ └── tests/ # 8-test validation suite (100% pass)
|
|
315
|
+
├── docs/ # Documentation
|
|
316
|
+
│ ├── user_guide.md # User manual
|
|
317
|
+
│ ├── tutorial.md # Step-by-step tutorials
|
|
318
|
+
│ └── api_reference.md # API documentation
|
|
319
|
+
├── modeling/ # Research/development (not in pip package)
|
|
320
|
+
├── notebooks/ # Example Jupyter notebooks
|
|
321
|
+
├── installation/ # Environment files
|
|
322
|
+
│ ├── microlive.yml # Conda env (macOS / CPU)
|
|
323
|
+
│ └── microlive_cuda.yml # Conda env (NVIDIA GPU)
|
|
324
|
+
├── tests/ # Test suite
|
|
325
|
+
├── pyproject.toml # Package configuration
|
|
326
|
+
└── LICENSE # GPL v3 License
|
|
327
|
+
```
|
|
328
|
+
|
|
329
|
+
---
|
|
330
|
+
|
|
331
|
+
## License
|
|
332
|
+
|
|
333
|
+
This project is licensed under the GNU General Public License v3 (GPLv3). See [LICENSE](LICENSE) for details.
|
|
334
|
+
|
|
335
|
+
---
|
|
336
|
+
|
|
337
|
+
## Citation
|
|
338
|
+
|
|
339
|
+
If you use MicroLive in your research, please cite:
|
|
340
|
+
|
|
341
|
+
> **Aguilera LU, Raymond WS, Sears RM, Nowling NL, Munsky B, Zhao N.** *MicroLive: An Image Processing Toolkit for Quantifying Live-cell Single-Molecule Microscopy.* bioRxiv, 2025.
|
|
342
|
+
> DOI: [10.1101/2025.09.25.678587](https://doi.org/10.1101/2025.09.25.678587)
|
|
343
|
+
|
|
344
|
+
```bibtex
|
|
345
|
+
@article{aguilera2025microlive,
|
|
346
|
+
title={MicroLive: An Image Processing Toolkit for Quantifying Live-cell Single-Molecule Microscopy},
|
|
347
|
+
author={Aguilera, Luis U and Raymond, William S and Sears, Rhiannon M and Nowling, Nathan L and Munsky, Brian and Zhao, Ning},
|
|
348
|
+
journal={bioRxiv},
|
|
349
|
+
pages={2025--09},
|
|
350
|
+
year={2025},
|
|
351
|
+
publisher={Cold Spring Harbor Laboratory},
|
|
352
|
+
doi={10.1101/2025.09.25.678587}
|
|
353
|
+
}
|
|
354
|
+
```
|
|
355
|
+
|
|
356
|
+
---
|
|
357
|
+
|
|
358
|
+
## Support
|
|
359
|
+
|
|
360
|
+
- **Documentation**: [User Guide](docs/user_guide.md) | [Tutorial](docs/tutorial.md) | [API Reference](docs/api_reference.md)
|
|
361
|
+
- **Issues & Contributions**: [GitHub](https://github.com/ningzhaoAnschutz/microlive/issues)
|
|
@@ -0,0 +1,26 @@
|
|
|
1
|
+
microlive/__init__.py,sha256=PKOH4vdUSLhXdjtrW-i0a_wtgIxb9K88tcXcUB1qOvY,1385
|
|
2
|
+
microlive/imports.py,sha256=VAAMavSLIKO0LooadTXfCdZiv8LQbV_wITeIv8IHwxM,7531
|
|
3
|
+
microlive/microscopy.py,sha256=D8mq7ssP7yw0tGCw2EESxH82LbGqYFKfVx3pDjky_pY,708504
|
|
4
|
+
microlive/ml_spot_detection.py,sha256=pVbOSGNJ0WWMuPRML42rFwvjKVZ0B1fJux1179OIbAg,10603
|
|
5
|
+
microlive/data/__init__.py,sha256=47DEQpj8HBSa-_TImW-5JCeuQeRkm5NMpJWZG3hSuFU,0
|
|
6
|
+
microlive/data/icons/__init__.py,sha256=47DEQpj8HBSa-_TImW-5JCeuQeRkm5NMpJWZG3hSuFU,0
|
|
7
|
+
microlive/data/icons/icon_micro.png,sha256=b5tFv4E6vUmLwYmYeM4PJuxLV_XqEzN14ueolekTFW0,370236
|
|
8
|
+
microlive/data/models/__init__.py,sha256=47DEQpj8HBSa-_TImW-5JCeuQeRkm5NMpJWZG3hSuFU,0
|
|
9
|
+
microlive/gui/__init__.py,sha256=tB-CdDC7x5OwYFAQxLOUvfVnUThaXKXVRsB68YP0Y6Q,28
|
|
10
|
+
microlive/gui/app.py,sha256=GTl2Iwe5uG603Ja6ykwfSG2kB7YaZJYQukLpC0DOurw,787890
|
|
11
|
+
microlive/gui/main.py,sha256=b66W_2V-pclGKOozfs75pwrCGbL_jkVU3kFt8RFMZIc,2520
|
|
12
|
+
microlive/gui/micro_mac.command,sha256=TkxYOO_5A2AiNJMz3_--1geBYfl77THpOLFZnV4J2ac,444
|
|
13
|
+
microlive/gui/micro_windows.bat,sha256=DJUKPhDbCO4HToLwSMT-QTYRe9Kr1wn5A2Ijy2klIrw,773
|
|
14
|
+
microlive/pipelines/__init__.py,sha256=VimchYrIWalFs_edRmjR1zBHIg2CcpRceZoRmB1e8kA,764
|
|
15
|
+
microlive/pipelines/pipeline_FRAP.py,sha256=GH5CqX6so1eWE07PXRCJZLAAqCzOqW-AoCqGXifapYE,56280
|
|
16
|
+
microlive/pipelines/pipeline_folding_efficiency.py,sha256=0PTogfXHRtO2kXOeQXb5-VBb46DQsj6namGVEkMGI0g,22550
|
|
17
|
+
microlive/pipelines/pipeline_particle_tracking.py,sha256=euPTLH6O9I66HkUb4Izah8ZF_aOdQLRyyR8vo1jSkFA,28245
|
|
18
|
+
microlive/pipelines/pipeline_spot_detection_no_tracking.py,sha256=t-p1xCQvThnVKMJZgk3Xhk3k6cvp1VgwTJ0ZIbfzNG0,19087
|
|
19
|
+
microlive/utils/__init__.py,sha256=5Ut2PeA0V5dM0VysmPpGH9OB-nmWDydzDkpRUwXfMHw,323
|
|
20
|
+
microlive/utils/device.py,sha256=tcPMU8UiXL-DuGwhudUgrbjW1lgIK_EUKIOeOn0U6q4,2533
|
|
21
|
+
microlive/utils/resources.py,sha256=Jz7kPI75xMLCBJMyX7Y_3ixKi_UgydfQkF0BlFtLCKs,1753
|
|
22
|
+
microlive-1.0.11.dist-info/METADATA,sha256=V5sp6sSjAp_JLMa1MLt-V7W4XXE3WbqZgqnJHtvsoO4,12432
|
|
23
|
+
microlive-1.0.11.dist-info/WHEEL,sha256=WLgqFyCfm_KASv4WHyYy0P3pM_m7J5L9k2skdKLirC8,87
|
|
24
|
+
microlive-1.0.11.dist-info/entry_points.txt,sha256=Zqp2vixyD8lngcfEmOi8fkCj7vPhesz5xlGBI-EubRw,54
|
|
25
|
+
microlive-1.0.11.dist-info/licenses/LICENSE,sha256=ixuiBLtpoK3iv89l7ylKkg9rs2GzF9ukPH7ynZYzK5s,35148
|
|
26
|
+
microlive-1.0.11.dist-info/RECORD,,
|