waveorder 2.2.1__py3-none-any.whl → 3.0.0__py3-none-any.whl
This diff represents the content of publicly available package versions that have been released to one of the supported registries. The information contained in this diff is provided for informational purposes only and reflects changes between package versions as they appear in their respective public registries.
- waveorder/_version.py +16 -3
- waveorder/acq/__init__.py +0 -0
- waveorder/acq/acq_functions.py +166 -0
- waveorder/assets/HSV_legend.png +0 -0
- waveorder/assets/JCh_legend.png +0 -0
- waveorder/assets/waveorder_plugin_logo.png +0 -0
- waveorder/calib/Calibration.py +1512 -0
- waveorder/calib/Optimization.py +470 -0
- waveorder/calib/__init__.py +0 -0
- waveorder/calib/calibration_workers.py +464 -0
- waveorder/cli/apply_inverse_models.py +328 -0
- waveorder/cli/apply_inverse_transfer_function.py +379 -0
- waveorder/cli/compute_transfer_function.py +432 -0
- waveorder/cli/gui_widget.py +58 -0
- waveorder/cli/main.py +39 -0
- waveorder/cli/monitor.py +163 -0
- waveorder/cli/option_eat_all.py +47 -0
- waveorder/cli/parsing.py +122 -0
- waveorder/cli/printing.py +16 -0
- waveorder/cli/reconstruct.py +67 -0
- waveorder/cli/settings.py +187 -0
- waveorder/cli/utils.py +175 -0
- waveorder/filter.py +1 -2
- waveorder/focus.py +136 -25
- waveorder/io/__init__.py +0 -0
- waveorder/io/_reader.py +61 -0
- waveorder/io/core_functions.py +272 -0
- waveorder/io/metadata_reader.py +195 -0
- waveorder/io/utils.py +175 -0
- waveorder/io/visualization.py +160 -0
- waveorder/models/inplane_oriented_thick_pol3d_vector.py +3 -3
- waveorder/models/isotropic_fluorescent_thick_3d.py +92 -0
- waveorder/models/isotropic_fluorescent_thin_3d.py +331 -0
- waveorder/models/isotropic_thin_3d.py +73 -72
- waveorder/models/phase_thick_3d.py +103 -4
- waveorder/napari.yaml +36 -0
- waveorder/plugin/__init__.py +9 -0
- waveorder/plugin/gui.py +1094 -0
- waveorder/plugin/gui.ui +1440 -0
- waveorder/plugin/job_manager.py +42 -0
- waveorder/plugin/main_widget.py +1605 -0
- waveorder/plugin/tab_recon.py +3294 -0
- waveorder/scripts/__init__.py +0 -0
- waveorder/scripts/launch_napari.py +13 -0
- waveorder/scripts/repeat-cal-acq-rec.py +147 -0
- waveorder/scripts/repeat-calibration.py +31 -0
- waveorder/scripts/samples.py +85 -0
- waveorder/scripts/simulate_zarr_acq.py +204 -0
- waveorder/util.py +1 -1
- waveorder/visuals/napari_visuals.py +1 -1
- waveorder-3.0.0.dist-info/METADATA +350 -0
- waveorder-3.0.0.dist-info/RECORD +69 -0
- {waveorder-2.2.1.dist-info → waveorder-3.0.0.dist-info}/WHEEL +1 -1
- waveorder-3.0.0.dist-info/entry_points.txt +5 -0
- {waveorder-2.2.1.dist-info → waveorder-3.0.0.dist-info}/licenses/LICENSE +13 -1
- waveorder-2.2.1.dist-info/METADATA +0 -188
- waveorder-2.2.1.dist-info/RECORD +0 -27
- {waveorder-2.2.1.dist-info → waveorder-3.0.0.dist-info}/top_level.txt +0 -0
|
@@ -0,0 +1,350 @@
|
|
|
1
|
+
Metadata-Version: 2.4
|
|
2
|
+
Name: waveorder
|
|
3
|
+
Version: 3.0.0
|
|
4
|
+
Summary: Wave-optical simulations and deconvolution of optical properties
|
|
5
|
+
Author-email: CZ Biohub SF <compmicro@czbiohub.org>
|
|
6
|
+
Maintainer-email: Talon Chandler <talon.chandler@czbiohub.org>, Shalin Mehta <shalin.mehta@czbiohub.org>
|
|
7
|
+
License: BSD 3-Clause License
|
|
8
|
+
|
|
9
|
+
Copyright (c) 2025, Chan Zuckerberg Biohub
|
|
10
|
+
|
|
11
|
+
Redistribution and use in source and binary forms, with or without
|
|
12
|
+
modification, are permitted provided that the following conditions are met:
|
|
13
|
+
|
|
14
|
+
1. Redistributions of source code must retain the above copyright notice, this
|
|
15
|
+
list of conditions and the following disclaimer.
|
|
16
|
+
|
|
17
|
+
2. Redistributions in binary form must reproduce the above copyright notice,
|
|
18
|
+
this list of conditions and the following disclaimer in the documentation
|
|
19
|
+
and/or other materials provided with the distribution.
|
|
20
|
+
|
|
21
|
+
3. Neither the name of the copyright holder nor the names of its
|
|
22
|
+
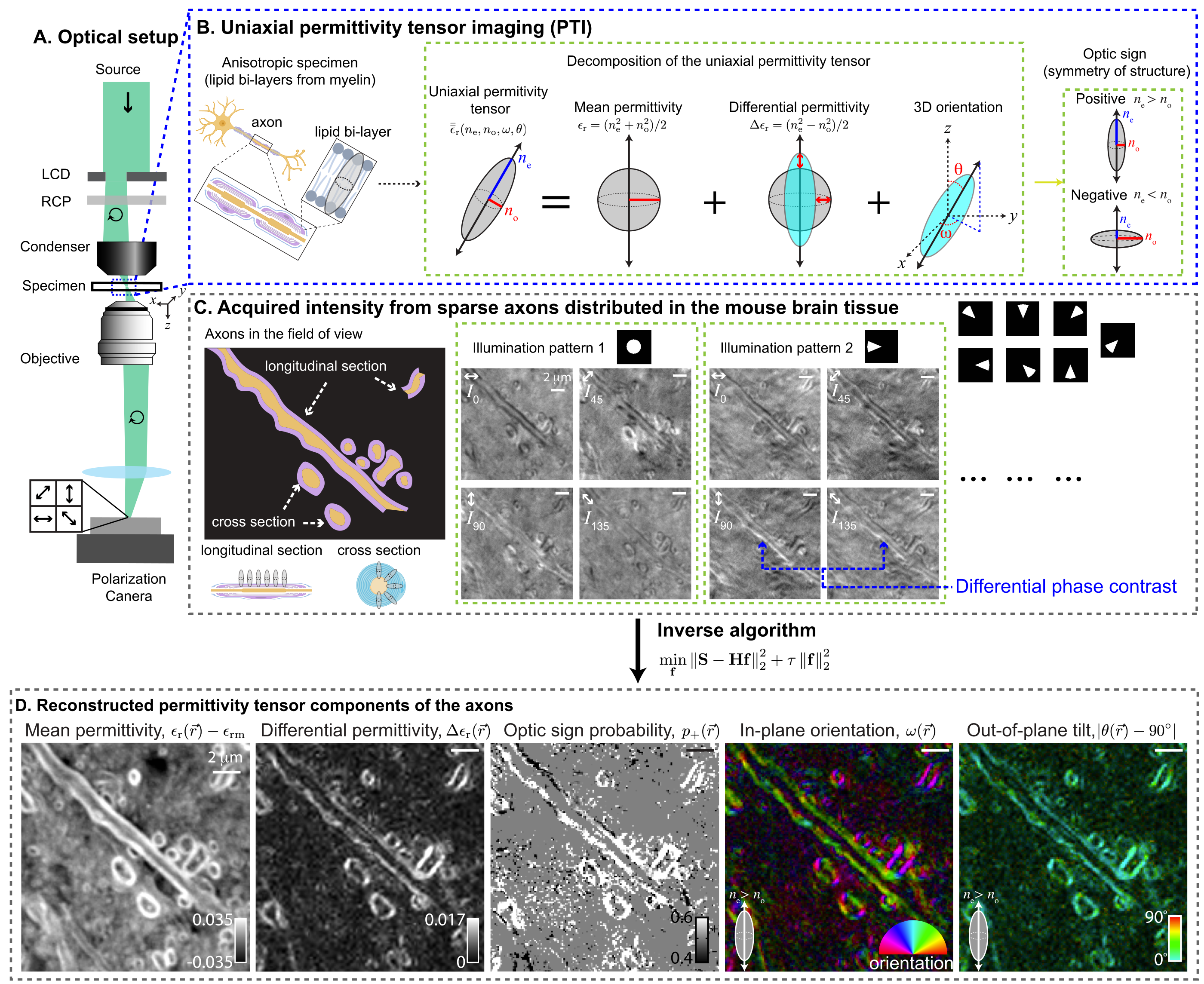
contributors may be used to endorse or promote products derived from
|
|
23
|
+
this software without specific prior written permission.
|
|
24
|
+
|
|
25
|
+
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
|
|
26
|
+
AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
|
|
27
|
+
IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
|
|
28
|
+
DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE
|
|
29
|
+
FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL
|
|
30
|
+
DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR
|
|
31
|
+
SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER
|
|
32
|
+
CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,
|
|
33
|
+
OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE
|
|
34
|
+
OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
|
|
35
|
+
|
|
36
|
+
## License Notice for Dependencies
|
|
37
|
+
|
|
38
|
+
This repository is licensed under the BSD License; however, it relies on certain
|
|
39
|
+
third-party dependencies that are licensed under the GNU General Public License
|
|
40
|
+
(GPL). Specifically:
|
|
41
|
+
|
|
42
|
+
- PyQt5 is licensed under the GNU General Public License v3 or later (GPLv3+).
|
|
43
|
+
This library is not included in this repository but must be installed separately
|
|
44
|
+
by users. Please be aware that the GPL license terms apply to these
|
|
45
|
+
dependencies, and certain uses of GPL-licensed code may have licensing
|
|
46
|
+
implications for your own software.
|
|
47
|
+
|
|
48
|
+
Project-URL: Homepage, https://github.com/mehta-lab/waveorder
|
|
49
|
+
Project-URL: Repository, https://github.com/mehta-lab/waveorder
|
|
50
|
+
Project-URL: Issues, https://github.com/mehta-lab/waveorder/issues
|
|
51
|
+
Keywords: simulation,optics,phase,scattering,polarization,label-free,permittivity,reconstruction-algorithm,qlipp,mipolscope,permittivity-tensor-imaging
|
|
52
|
+
Classifier: Development Status :: 4 - Beta
|
|
53
|
+
Classifier: Intended Audience :: Science/Research
|
|
54
|
+
Classifier: License :: OSI Approved :: BSD License
|
|
55
|
+
Classifier: Programming Language :: Python :: 3
|
|
56
|
+
Classifier: Programming Language :: Python :: 3.11
|
|
57
|
+
Classifier: Programming Language :: Python :: 3.12
|
|
58
|
+
Classifier: Programming Language :: Python :: 3.13
|
|
59
|
+
Classifier: Topic :: Scientific/Engineering
|
|
60
|
+
Classifier: Topic :: Scientific/Engineering :: Image Processing
|
|
61
|
+
Classifier: Topic :: Scientific/Engineering :: Bio-Informatics
|
|
62
|
+
Classifier: Operating System :: POSIX :: Linux
|
|
63
|
+
Classifier: Operating System :: Microsoft :: Windows
|
|
64
|
+
Classifier: Operating System :: MacOS
|
|
65
|
+
Requires-Python: >=3.11
|
|
66
|
+
Description-Content-Type: text/markdown
|
|
67
|
+
License-File: LICENSE
|
|
68
|
+
Requires-Dist: click>=8.0.1
|
|
69
|
+
Requires-Dist: colorspacious>=1.1.2
|
|
70
|
+
Requires-Dist: importlib-metadata
|
|
71
|
+
Requires-Dist: iohub<0.4,>=0.2
|
|
72
|
+
Requires-Dist: ipywidgets>=7.5.1
|
|
73
|
+
Requires-Dist: matplotlib>=3.1.1
|
|
74
|
+
Requires-Dist: natsort>=7.1.1
|
|
75
|
+
Requires-Dist: numpy>=1.24
|
|
76
|
+
Requires-Dist: psutil
|
|
77
|
+
Requires-Dist: pyqtgraph>=0.12.3
|
|
78
|
+
Requires-Dist: pydantic
|
|
79
|
+
Requires-Dist: pywavelets>=1.1.1
|
|
80
|
+
Requires-Dist: scipy>=1.3.0
|
|
81
|
+
Requires-Dist: torch>=2.4.1
|
|
82
|
+
Requires-Dist: qtpy
|
|
83
|
+
Requires-Dist: wget>=3.2
|
|
84
|
+
Provides-Extra: visual
|
|
85
|
+
Requires-Dist: napari[pyqt6]; extra == "visual"
|
|
86
|
+
Requires-Dist: PyQt6<6.8,>=6.5; extra == "visual"
|
|
87
|
+
Requires-Dist: napari-ome-zarr>=0.3.2; extra == "visual"
|
|
88
|
+
Requires-Dist: pycromanager==0.27.2; extra == "visual"
|
|
89
|
+
Requires-Dist: jupyter; extra == "visual"
|
|
90
|
+
Requires-Dist: jupytext; extra == "visual"
|
|
91
|
+
Provides-Extra: dev
|
|
92
|
+
Requires-Dist: black==25.1.0; extra == "dev"
|
|
93
|
+
Requires-Dist: click>=8.2.0; extra == "dev"
|
|
94
|
+
Requires-Dist: hypothesis; extra == "dev"
|
|
95
|
+
Requires-Dist: napari[pyqt6]; extra == "dev"
|
|
96
|
+
Requires-Dist: PyQt6<6.8,>=6.5; extra == "dev"
|
|
97
|
+
Requires-Dist: pre-commit; extra == "dev"
|
|
98
|
+
Requires-Dist: pytest-cov; extra == "dev"
|
|
99
|
+
Requires-Dist: pytest-qt; extra == "dev"
|
|
100
|
+
Requires-Dist: pytest>=5.0.0; extra == "dev"
|
|
101
|
+
Provides-Extra: docs
|
|
102
|
+
Requires-Dist: sphinx_copybutton; extra == "docs"
|
|
103
|
+
Requires-Dist: sphinxcontrib-video; extra == "docs"
|
|
104
|
+
Requires-Dist: myst_parser; extra == "docs"
|
|
105
|
+
Requires-Dist: sphinx_mdinclude; extra == "docs"
|
|
106
|
+
Requires-Dist: linkify-it-py; extra == "docs"
|
|
107
|
+
Requires-Dist: numpydoc; extra == "docs"
|
|
108
|
+
Requires-Dist: sphinx_sitemap; extra == "docs"
|
|
109
|
+
Requires-Dist: sphinx_gallery; extra == "docs"
|
|
110
|
+
Requires-Dist: pydata_sphinx_theme; extra == "docs"
|
|
111
|
+
Requires-Dist: matplotlib; extra == "docs"
|
|
112
|
+
Requires-Dist: importlib_metadata; extra == "docs"
|
|
113
|
+
Provides-Extra: all
|
|
114
|
+
Requires-Dist: waveorder[visual]; extra == "all"
|
|
115
|
+
Requires-Dist: waveorder[dev]; extra == "all"
|
|
116
|
+
Requires-Dist: waveorder[docs]; extra == "all"
|
|
117
|
+
Dynamic: license-file
|
|
118
|
+
|
|
119
|
+
<p align="center">
|
|
120
|
+
<img src="https://github.com/user-attachments/assets/00367e51-0e0a-445e-8d12-2a273f9014aa" alt="Image" width="50%">
|
|
121
|
+
</p>
|
|
122
|
+
|
|
123
|
+
[](https://pypi.org/project/waveorder)
|
|
124
|
+
[](https://pypistats.org/packages/waveorder)
|
|
125
|
+
[](https://pepy.tech/project/waveorder)
|
|
126
|
+
[](https://github.com/mehta-lab/waveorder/graphs/contributors)
|
|
127
|
+

|
|
128
|
+

|
|
129
|
+

|
|
130
|
+
|
|
131
|
+
Label-agnostic computational microscopy of architectural order.
|
|
132
|
+
|
|
133
|
+
# Overview
|
|
134
|
+
|
|
135
|
+
`waveorder` is a generalist framework for label-agnostic computational microscopy of architectural order, i.e., density, alignment, and orientation of biomolecules with the spatial resolution down to the diffraction limit. The framework implements wave-optical simulations and corresponding reconstruction algorithms for diverse label-free and fluorescence computational imaging methods that enable quantitative imaging of the architecture of dynamic cell systems.
|
|
136
|
+
|
|
137
|
+
Our goal is to enable modular and user-friendly implementations of computational microscopy methods for dynamic imaging across the scales of organelles, cells, and tissues.
|
|
138
|
+
|
|
139
|
+
|
|
140
|
+
The framework is described in the following [preprint](https://arxiv.org/abs/2412.09775).
|
|
141
|
+
|
|
142
|
+
https://github.com/user-attachments/assets/4f9969e5-94ce-4e08-9f30-68314a905db6
|
|
143
|
+
|
|
144
|
+
<details>
|
|
145
|
+
`waveorder` enables simulations and reconstructions of label-agnostic microscopy data as described in the following [preprint](https://arxiv.org/abs/2412.09775)
|
|
146
|
+
<summary> Chandler et al. 2024 </summary>
|
|
147
|
+
<pre><code>
|
|
148
|
+
@article{chandler_2024,
|
|
149
|
+
author = {Chandler, Talon and Hirata-Miyasaki, Eduardo and Ivanov, Ivan E. and Liu, Ziwen and Sundarraman, Deepika and Ryan, Allyson Quinn and Jacobo, Adrian and Balla, Keir and Mehta, Shalin B.},
|
|
150
|
+
title = {waveOrder: generalist framework for label-agnostic computational microscopy},
|
|
151
|
+
journal = {arXiv},
|
|
152
|
+
year = {2024},
|
|
153
|
+
month = dec,
|
|
154
|
+
eprint = {2412.09775},
|
|
155
|
+
doi = {10.48550/arXiv.2412.09775}
|
|
156
|
+
}
|
|
157
|
+
</code></pre>
|
|
158
|
+
</details>
|
|
159
|
+
|
|
160
|
+
# Computational Microscopy Methods
|
|
161
|
+
|
|
162
|
+
`waveorder` framework enables simulations and reconstructions of data for diverse one-photon (single-scattering based) computational microscopy methods, summarized below.
|
|
163
|
+
|
|
164
|
+
## Label-free microscopy
|
|
165
|
+
|
|
166
|
+
### Quantitative label-free imaging with phase and polarization (QLIPP)
|
|
167
|
+
|
|
168
|
+
Acquisition, calibration, background correction, reconstruction, and applications of QLIPP are described in the following [E-Life Paper](https://elifesciences.org/articles/55502):
|
|
169
|
+
|
|
170
|
+
[](https://www.youtube.com/watch?v=JEZAaPeZhck)
|
|
171
|
+
|
|
172
|
+
<details>
|
|
173
|
+
<summary> Guo et al. 2020 </summary>
|
|
174
|
+
<pre><code>
|
|
175
|
+
@article{guo_2020,
|
|
176
|
+
author = {Guo, Syuan-Ming and Yeh, Li-Hao and Folkesson, Jenny and Ivanov, Ivan E. and Krishnan, Anitha P. and Keefe, Matthew G. and Hashemi, Ezzat and Shin, David and Chhun, Bryant B. and Cho, Nathan H. and Leonetti, Manuel D. and Han, May H. and Nowakowski, Tomasz J. and Mehta, Shalin B.},
|
|
177
|
+
title = {Revealing architectural order with quantitative label-free imaging and deep learning},
|
|
178
|
+
journal = {eLife},
|
|
179
|
+
volume = {9},
|
|
180
|
+
pages = {e55502},
|
|
181
|
+
year = {2020},
|
|
182
|
+
doi = {10.7554/eLife.55502}
|
|
183
|
+
}
|
|
184
|
+
</code></pre>
|
|
185
|
+
</details>
|
|
186
|
+
|
|
187
|
+
### Permittivity tensor imaging (PTI)
|
|
188
|
+
|
|
189
|
+
PTI provides volumetric reconstructions of mean permittivity ($\propto$ material density), differential permittivity ($\propto$ material anisotropy), 3D orientation, and optic sign. The following figure summarizes PTI acquisition and reconstruction with a small optical section of the mouse brain tissue:
|
|
190
|
+
|
|
191
|
+
|
|
192
|
+
|
|
193
|
+
The acquisition, calibration, background correction, reconstruction, and applications of PTI are described in the following [paper](https://doi.org/10.1101/2020.12.15.422951) published in Nature Methods:
|
|
194
|
+
|
|
195
|
+
<details>
|
|
196
|
+
<summary> Yeh et al. 2024 </summary>
|
|
197
|
+
<pre><code>
|
|
198
|
+
@article{yeh_2024,
|
|
199
|
+
author = {Yeh, Li-Hao and Ivanov, Ivan E. and Chandler, Talon and Byrum, Janie R. and Chhun, Bryant B. and Guo, Syuan-Ming and Foltz, Cameron and Hashemi, Ezzat and Perez-Bermejo, Juan A. and Wang, Huijun and Yu, Yanhao and Kazansky, Peter G. and Conklin, Bruce R. and Han, May H. and Mehta, Shalin B.},
|
|
200
|
+
title = {Permittivity tensor imaging: modular label-free imaging of 3D dry mass and 3D orientation at high resolution},
|
|
201
|
+
journal = {Nature Methods},
|
|
202
|
+
volume = {21},
|
|
203
|
+
number = {7},
|
|
204
|
+
pages = {1257--1274},
|
|
205
|
+
year = {2024},
|
|
206
|
+
month = jul,
|
|
207
|
+
issn = {1548-7105},
|
|
208
|
+
publisher = {Nature Publishing Group},
|
|
209
|
+
doi = {10.1038/s41592-024-02291-w}
|
|
210
|
+
}
|
|
211
|
+
</code></pre>
|
|
212
|
+
</details>
|
|
213
|
+
|
|
214
|
+
|
|
215
|
+
### Quantitative phase imaging (QPI) from defocus
|
|
216
|
+
__phase__ from a volumetric brightfield acquisition ([2D phase](https://www.osapublishing.org/ao/abstract.cfm?uri=ao-54-28-8566)/[3D phase](https://www.osapublishing.org/ao/abstract.cfm?uri=ao-57-1-a205))
|
|
217
|
+
|
|
218
|
+

|
|
219
|
+
|
|
220
|
+
|
|
221
|
+
<details>
|
|
222
|
+
<summary> Jenkins and Gaylord 2015 (2D QPI from defocus) </summary>
|
|
223
|
+
<pre><code>
|
|
224
|
+
@article{Jenkins:15,
|
|
225
|
+
author = {Micah H. Jenkins and Thomas K. Gaylord},
|
|
226
|
+
journal = {Appl. Opt.},
|
|
227
|
+
keywords = {Phase retrieval; Partial coherence in imaging; Interferometric imaging ; Imaging systems; Microlens arrays; Optical transfer functions; Phase contrast; Spatial resolution; Three dimensional imaging},
|
|
228
|
+
number = {28},
|
|
229
|
+
pages = {8566--8579},
|
|
230
|
+
publisher = {Optica Publishing Group},
|
|
231
|
+
title = {Quantitative phase microscopy via optimized inversion of the phase optical transfer function},
|
|
232
|
+
volume = {54},
|
|
233
|
+
month = {Oct},
|
|
234
|
+
year = {2015},
|
|
235
|
+
url = {https://opg.optica.org/ao/abstract.cfm?URI=ao-54-28-8566},
|
|
236
|
+
doi = {10.1364/AO.54.008566},
|
|
237
|
+
}
|
|
238
|
+
</code></pre>
|
|
239
|
+
</details>
|
|
240
|
+
|
|
241
|
+
<details>
|
|
242
|
+
<summary> Soto, Rodrigo, and Alieva 2018 (3D QPI from defocus) </summary>
|
|
243
|
+
<pre><code>
|
|
244
|
+
@article{Soto:18,
|
|
245
|
+
author = {Juan M. Soto and Jos\'{e} A. Rodrigo and Tatiana Alieva},
|
|
246
|
+
journal = {Appl. Opt.},
|
|
247
|
+
keywords = {Coherence and statistical optics; Image reconstruction techniques; Optical transfer functions; Optical inspection; Three-dimensional microscopy; Acoustooptic modulators; Illumination design; Inverse design; LED sources; Three dimensional imaging; Three dimensional reconstruction},
|
|
248
|
+
number = {1},
|
|
249
|
+
pages = {A205--A214},
|
|
250
|
+
publisher = {Optica Publishing Group},
|
|
251
|
+
title = {Optical diffraction tomography with fully and partially coherent illumination in high numerical aperture label-free microscopy \[Invited\]},
|
|
252
|
+
volume = {57},
|
|
253
|
+
month = {Jan},
|
|
254
|
+
year = {2018},
|
|
255
|
+
url = {https://opg.optica.org/ao/abstract.cfm?URI=ao-57-1-A205},
|
|
256
|
+
doi = {10.1364/AO.57.00A205},
|
|
257
|
+
}
|
|
258
|
+
</code></pre>
|
|
259
|
+
</details>
|
|
260
|
+
|
|
261
|
+
### QPI with differential phase contrast
|
|
262
|
+
__phase__ from differential phase contrast
|
|
263
|
+
|
|
264
|
+
***Work in progress***
|
|
265
|
+
|
|
266
|
+
* [2D](https://www.osapublishing.org/oe/fulltext.cfm?uri=oe-23-9-11394&id=315599) DPC
|
|
267
|
+
* [3D](https://www.osapublishing.org/boe/fulltext.cfm?uri=boe-7-10-3940&id=349951) DPC
|
|
268
|
+
|
|
269
|
+
|
|
270
|
+
## Fluorescence microscopy
|
|
271
|
+
|
|
272
|
+
### Widefield deconvolution microscopy
|
|
273
|
+
__fluorescence density__ from a widefield volumetric fluorescence acquisition.
|
|
274
|
+
|
|
275
|
+
<details>
|
|
276
|
+
<summary> Swedlow 2013 </summary>
|
|
277
|
+
<pre><code>
|
|
278
|
+
@article{Swedlow:13,
|
|
279
|
+
author = {Swedlow, John R.},
|
|
280
|
+
journal = {Methods Cell Biol.},
|
|
281
|
+
title = {Quantitative fluorescence microscopy and image deconvolution},
|
|
282
|
+
year = {2013},
|
|
283
|
+
volume = {114},
|
|
284
|
+
pages = {407--26},
|
|
285
|
+
doi = {10.1016/B978-0-12-407761-4.00017-8}
|
|
286
|
+
}
|
|
287
|
+
</code></pre>
|
|
288
|
+
</details>
|
|
289
|
+
|
|
290
|
+
### Oblique plane light-sheet microscopy
|
|
291
|
+
__fluorescence density__ from oblique plane light-sheet microscopy.
|
|
292
|
+
|
|
293
|
+
<details>
|
|
294
|
+
<summary> Ivanov, Hirata-Miyasaki, Chandler et al. 2024 </summary>
|
|
295
|
+
<pre><code>
|
|
296
|
+
@article{ivanov_2024,
|
|
297
|
+
author = {Ivanov, Ivan E. and Hirata-Miyasaki, Eduardo and Chandler, Talon and Cheloor-Kovilakam, Rasmi and Liu, Ziwen and Pradeep, Soorya and Liu, Chad and Bhave, Madhura and Khadka, Sudip and Arias, Carolina and Leonetti, Manuel D. and Huang, Bo and Mehta, Shalin B.},
|
|
298
|
+
title = {Mantis: High-throughput 4D imaging and analysis of the molecular and physical architecture of cells},
|
|
299
|
+
journal = {PNAS Nexus},
|
|
300
|
+
volume = {3},
|
|
301
|
+
number = {9},
|
|
302
|
+
pages = {pgae323},
|
|
303
|
+
year = {2024},
|
|
304
|
+
doi = {10.1093/pnasnexus/pgae323}
|
|
305
|
+
</code></pre>
|
|
306
|
+
</details>
|
|
307
|
+
|
|
308
|
+
|
|
309
|
+
## Citation
|
|
310
|
+
|
|
311
|
+
Please cite this repository, along with the relevant publications and preprints, if you use or adapt this code. The citation information can be found by clicking "Cite this repository" button in the About section in the right sidebar.
|
|
312
|
+
|
|
313
|
+
## Installation
|
|
314
|
+
|
|
315
|
+
Create a virtual environment:
|
|
316
|
+
|
|
317
|
+
```sh
|
|
318
|
+
conda create -y -n waveorder python=3.12
|
|
319
|
+
conda activate waveorder
|
|
320
|
+
```
|
|
321
|
+
|
|
322
|
+
Most users should install `waveorder` with:
|
|
323
|
+
```sh
|
|
324
|
+
pip install waveorder[visual]
|
|
325
|
+
```
|
|
326
|
+
|
|
327
|
+
We also maintain three dependency sets for different interfaces:
|
|
328
|
+
|
|
329
|
+
```sh
|
|
330
|
+
pip install waveorder # API, CLI
|
|
331
|
+
pip install waveorder[visual] # API, CLI, GUI
|
|
332
|
+
pip install waveorder[all] # API, CLI, GUI, docs, dev dependencies
|
|
333
|
+
```
|
|
334
|
+
|
|
335
|
+
|
|
336
|
+
|
|
337
|
+
(M1 users) `pytorch` has [incomplete GPU support](https://github.com/pytorch/pytorch/issues/77764),
|
|
338
|
+
so please use `export PYTORCH_ENABLE_MPS_FALLBACK=1`
|
|
339
|
+
to allow some operators to fallback to CPU if you plan to use GPU acceleration for polarization reconstruction.
|
|
340
|
+
|
|
341
|
+
|
|
342
|
+
## Examples
|
|
343
|
+
The [examples](https://github.com/mehta-lab/waveorder/tree/main/docs/examples) illustrate simulations and reconstruction for 2D QLIPP, 3D phase from brightfield, and 2D/3D PTI methods. To run examples, use
|
|
344
|
+
|
|
345
|
+
```sh
|
|
346
|
+
git clone https://github.com/mehta-lab/waveorder.git
|
|
347
|
+
cd waveorder
|
|
348
|
+
pip install -e .[all]
|
|
349
|
+
python ./docs/examples/models/phase_thick_3d.py
|
|
350
|
+
```
|
|
@@ -0,0 +1,69 @@
|
|
|
1
|
+
waveorder/__init__.py,sha256=47DEQpj8HBSa-_TImW-5JCeuQeRkm5NMpJWZG3hSuFU,0
|
|
2
|
+
waveorder/_version.py,sha256=9fL11DDeXfhKmxyz_HrYU3Yy7BjMWrJklCddPlmAj6A,704
|
|
3
|
+
waveorder/background_estimator.py,sha256=GFy3N0qqp5M6JVZBbIUvTYhSin8Njg7zA2WN69pKLAE,12398
|
|
4
|
+
waveorder/correction.py,sha256=uAWDKXq-FYwi1obxxWq0A-suNVf1cvqUnPsDC-LIlsM,3460
|
|
5
|
+
waveorder/filter.py,sha256=UatcxE3xqwwB1p0IVoEjH8Qw0CB6hvcXvnRR6IRNXsQ,7325
|
|
6
|
+
waveorder/focus.py,sha256=4IenjsMfwDkZlSImNMB3mnyuDz4XF7VJWiaCmzDLseI,10928
|
|
7
|
+
waveorder/napari.yaml,sha256=iqmzGwSopu-L38yxuAnhMTuBvDqKIm2mTWfxjn6Duks,1463
|
|
8
|
+
waveorder/optics.py,sha256=RikR3kkPFoVUS86jiGQZey7Jtz5Ga-xKWnXsexo3Okc,44242
|
|
9
|
+
waveorder/reconstruct.py,sha256=nF00-uxYF67QVcNClp1VIB31ElUfbjO3RNxbxc0ELH4,832
|
|
10
|
+
waveorder/sampling.py,sha256=OAqlfjEemX6tQV2a2S8X0wQx9JCSykcvVxvKr1CY3H0,2521
|
|
11
|
+
waveorder/stokes.py,sha256=G_hcqS-GbslVnCWmS13y77K1YZ8cvwIl-DDKyC84LSI,15184
|
|
12
|
+
waveorder/util.py,sha256=axo2XvguW2UGb6IIXzgZVlP2IDxP4qUT_hi3DUTUwv8,71236
|
|
13
|
+
waveorder/waveorder_reconstructor.py,sha256=-MluWmnZnCZm7Xu-8V8QWX9ma0_5oAZO2Xlyrg1bRes,152072
|
|
14
|
+
waveorder/waveorder_simulator.py,sha256=uRRX_wcWzJzlVcfToLpIlh4e8Xt9NjTvdonyGEf2Z1c,45805
|
|
15
|
+
waveorder/acq/__init__.py,sha256=47DEQpj8HBSa-_TImW-5JCeuQeRkm5NMpJWZG3hSuFU,0
|
|
16
|
+
waveorder/acq/acq_functions.py,sha256=4GJCkxncFunsRbLesf1LErY7CaA26dMh2goVVUr72y0,5128
|
|
17
|
+
waveorder/assets/HSV_legend.png,sha256=7k9l23V-is7TIVnKchIat9JE_UGSqjbRzgVROeNrNJU,12132
|
|
18
|
+
waveorder/assets/JCh_legend.png,sha256=waQ980-7-APUqzuvBjKh5PKE91jaCbKire0NJf-y6AQ,14900
|
|
19
|
+
waveorder/assets/waveorder_plugin_logo.png,sha256=3advwUtIqhXJmHrqXF3NmWB8OMjQP2N-Nsbi-IaIQw8,14686
|
|
20
|
+
waveorder/calib/Calibration.py,sha256=Fl6zQOJg4V6nKtbAlNL7s9kslmrxk6q-LAueh59F1bA,52007
|
|
21
|
+
waveorder/calib/Optimization.py,sha256=uBQRvZ3tsG52pOSwnRj3Lduf_wVTTYfcjegAZsN58UE,13909
|
|
22
|
+
waveorder/calib/__init__.py,sha256=47DEQpj8HBSa-_TImW-5JCeuQeRkm5NMpJWZG3hSuFU,0
|
|
23
|
+
waveorder/calib/calibration_workers.py,sha256=xI1Zg_D6m1wuls4af0Nlzl_3fADeAeHzSO2gWKGaORo,15325
|
|
24
|
+
waveorder/cli/apply_inverse_models.py,sha256=8nrwj4bgHnha59uOvvV0PqdRyjyldWfUs_ZbfdtJ-ZE,10344
|
|
25
|
+
waveorder/cli/apply_inverse_transfer_function.py,sha256=EgmzIoDX4o7GqYIQfk7wgJZ8DxqfXeTAJjDXbMOqWPY,13369
|
|
26
|
+
waveorder/cli/compute_transfer_function.py,sha256=IfOtxVSYqcF9urDBxDftm8Txw3h6jNzAswIOgnXtRf0,14319
|
|
27
|
+
waveorder/cli/gui_widget.py,sha256=bPbeAAQEFzD2WnlO_vKZ5RSdaFi6J3Psouzfuj7u9f4,1343
|
|
28
|
+
waveorder/cli/main.py,sha256=uW18kwZOpYbgUKfGXexIF3LITxkUJeRMHnTmAwAPWjc,942
|
|
29
|
+
waveorder/cli/monitor.py,sha256=XBhqyd7dxnXrfca23rm0oAILEnx8lEVPsS2oSfmt8kw,4757
|
|
30
|
+
waveorder/cli/option_eat_all.py,sha256=GJgI4owf59IhZTtQiLsUy19WQJLJDXB-Gg7WIf5aREw,1823
|
|
31
|
+
waveorder/cli/parsing.py,sha256=IWSjRCp2KQiEzw4T_aCvMSoPrKoRRbIJP0Tc8Pcb-R4,3476
|
|
32
|
+
waveorder/cli/printing.py,sha256=tCWRE7YnE4ByROAhaYlCjJLnCwZ3IEcm3Bo6ZoO1uxI,326
|
|
33
|
+
waveorder/cli/reconstruct.py,sha256=qbFCoMZC9utDofx8oU0BUljMC-MoN_ykrp1sXGe39zo,1792
|
|
34
|
+
waveorder/cli/settings.py,sha256=d5EmqnzNjwDHVnP4xONAamBe6I4rdNHWS1pUXKyu1bw,6604
|
|
35
|
+
waveorder/cli/utils.py,sha256=-7RYPUEzhTPhmgQUSptNxE1GmnAjzuucixEBWcRL0KI,5667
|
|
36
|
+
waveorder/io/__init__.py,sha256=47DEQpj8HBSa-_TImW-5JCeuQeRkm5NMpJWZG3hSuFU,0
|
|
37
|
+
waveorder/io/_reader.py,sha256=l17s_-1aARCFn76VIpDljoOIL088uPx3A1mTl3T5l9o,1630
|
|
38
|
+
waveorder/io/core_functions.py,sha256=g4HrkfcQinEhCPdt0FtRSXRXUjRL1ZQe5g0sClnjVZA,6553
|
|
39
|
+
waveorder/io/metadata_reader.py,sha256=DiVqdJiwSduHa6eOrfJfcky1ZoFI3ySPEXJrbuFM8fg,5483
|
|
40
|
+
waveorder/io/utils.py,sha256=ArmKCq5_Wj3tyiD9Zl6jaUkjgIslJffJj5xRUGtMcMM,4804
|
|
41
|
+
waveorder/io/visualization.py,sha256=u_EGSc2s1pkB-xvGVxmmMr2zQObihjkV0PCB_7yizak,4753
|
|
42
|
+
waveorder/models/inplane_oriented_thick_pol3d.py,sha256=bx5yViNz7wY5BBXUea1Tw0RhYsEzErENRnAbgpa34C0,5992
|
|
43
|
+
waveorder/models/inplane_oriented_thick_pol3d_vector.py,sha256=Qh1E0fWHh6KThQnW3TYBGmDtNrO98n9W42t6G_nQO5w,9759
|
|
44
|
+
waveorder/models/isotropic_fluorescent_thick_3d.py,sha256=0bu9OdYj2aP1phzRxod8tExURaYdgwddAD61BbGF-lg,10387
|
|
45
|
+
waveorder/models/isotropic_fluorescent_thin_3d.py,sha256=uOxRTOKu5Viv_KTKlW7c91M7hREx3eER-owKFyrhRCQ,10239
|
|
46
|
+
waveorder/models/isotropic_thin_3d.py,sha256=BPp0DR3Pu8b7NpnGXUJ1ZqdGbs_P7-H5iVGagVClPtg,11328
|
|
47
|
+
waveorder/models/phase_thick_3d.py,sha256=eUmf_AbSDMwFNDfE8WPGr5sJShvL4YAjsZff_KO559k,12323
|
|
48
|
+
waveorder/plugin/__init__.py,sha256=4Idw8HLl6LZ-E0cgW9mSsCAQk-D4Hnmoi0W9iaV6yrw,333
|
|
49
|
+
waveorder/plugin/gui.py,sha256=NazopP-wb3Fup4FWfgH4zWdkOKvilpxWrOXgsI0xquI,50593
|
|
50
|
+
waveorder/plugin/gui.ui,sha256=7_CUDI2Wqbh2EBal9-ZmNJcb7cCCxiZcQe-DO6NBnM4,49920
|
|
51
|
+
waveorder/plugin/job_manager.py,sha256=FNQ9T-djbJM2SzRx99khybCjEF5vG-8pS6jEBHqo7og,1157
|
|
52
|
+
waveorder/plugin/main_widget.py,sha256=Vf5Eve9ik_86mdsSbezacME96eFnDf7A9myDaCnHHt4,58098
|
|
53
|
+
waveorder/plugin/tab_recon.py,sha256=kiMhTa6sxqmQAYw9gGpcu66y0vEB-_e5mMAP7V136qU,131698
|
|
54
|
+
waveorder/scripts/__init__.py,sha256=47DEQpj8HBSa-_TImW-5JCeuQeRkm5NMpJWZG3hSuFU,0
|
|
55
|
+
waveorder/scripts/launch_napari.py,sha256=0Sx_2XCJ38pBfe7QvthUn0Vu8ZcqD6qHjwBvzR6V2JY,221
|
|
56
|
+
waveorder/scripts/repeat-cal-acq-rec.py,sha256=XVdQSoTacNczd58MBty4Qa4umprdUkgiwCkWQQr7p4g,3908
|
|
57
|
+
waveorder/scripts/repeat-calibration.py,sha256=mHLM-axQh2h8CE5F11-qfGLoZoHCKOh8IWThESqKgRI,725
|
|
58
|
+
waveorder/scripts/samples.py,sha256=n5PEv2mLXmuuqYhjtzxu7SDeW3MJJgzoYPrGfnX_Poo,3076
|
|
59
|
+
waveorder/scripts/simulate_zarr_acq.py,sha256=AGVKNsi46J0AyDJvgSCOMQqtOIhcNWY5yCMgyeCQTZw,6390
|
|
60
|
+
waveorder/visuals/jupyter_visuals.py,sha256=6kxICjEtP1qc1EuETc_NJ6Y4A7nVaC-bP3wl_CQNPfg,58096
|
|
61
|
+
waveorder/visuals/matplotlib_visuals.py,sha256=v1zi0ZlXEV5CcpNzTWL0vDJ2Md2-RSHnc3rAB61wimg,10915
|
|
62
|
+
waveorder/visuals/napari_visuals.py,sha256=bDEgHvoexdsGW4pIbnLfBCU4e4be41PMyKx9Mq9so1c,2361
|
|
63
|
+
waveorder/visuals/utils.py,sha256=QC5WSc2yzPMjk66IjA39iNFgO_0It6evma201hH8Lg4,1001
|
|
64
|
+
waveorder-3.0.0.dist-info/licenses/LICENSE,sha256=MATRcU1PWMarfKd4-d4q6eMdy4JWzC6fpavJI_zXHeE,2068
|
|
65
|
+
waveorder-3.0.0.dist-info/METADATA,sha256=rn2NzZgz2ev6HQupT9Y_M8-aRNU67ncoFTnx7FBHZBQ,15574
|
|
66
|
+
waveorder-3.0.0.dist-info/WHEEL,sha256=wUyA8OaulRlbfwMtmQsvNngGrxQHAvkKcvRmdizlJi0,92
|
|
67
|
+
waveorder-3.0.0.dist-info/entry_points.txt,sha256=cOCI4v8cdinVJiyAMlYZtxK6GCS4GF6Qpk1hTsgW18Y,106
|
|
68
|
+
waveorder-3.0.0.dist-info/top_level.txt,sha256=i3zReXiiMTnyPk93W7aEz_oEfsLnfR_Kzl7PW7kUslA,10
|
|
69
|
+
waveorder-3.0.0.dist-info/RECORD,,
|
|
@@ -1,6 +1,6 @@
|
|
|
1
1
|
BSD 3-Clause License
|
|
2
2
|
|
|
3
|
-
Copyright (c)
|
|
3
|
+
Copyright (c) 2025, Chan Zuckerberg Biohub
|
|
4
4
|
|
|
5
5
|
Redistribution and use in source and binary forms, with or without
|
|
6
6
|
modification, are permitted provided that the following conditions are met:
|
|
@@ -26,3 +26,15 @@ SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER
|
|
|
26
26
|
CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,
|
|
27
27
|
OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE
|
|
28
28
|
OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
|
|
29
|
+
|
|
30
|
+
## License Notice for Dependencies
|
|
31
|
+
|
|
32
|
+
This repository is licensed under the BSD License; however, it relies on certain
|
|
33
|
+
third-party dependencies that are licensed under the GNU General Public License
|
|
34
|
+
(GPL). Specifically:
|
|
35
|
+
|
|
36
|
+
- PyQt5 is licensed under the GNU General Public License v3 or later (GPLv3+).
|
|
37
|
+
This library is not included in this repository but must be installed separately
|
|
38
|
+
by users. Please be aware that the GPL license terms apply to these
|
|
39
|
+
dependencies, and certain uses of GPL-licensed code may have licensing
|
|
40
|
+
implications for your own software.
|
|
@@ -1,188 +0,0 @@
|
|
|
1
|
-
Metadata-Version: 2.4
|
|
2
|
-
Name: waveorder
|
|
3
|
-
Version: 2.2.1
|
|
4
|
-
Summary: Wave-optical simulations and deconvolution of optical properties
|
|
5
|
-
Author-email: CZ Biohub SF <compmicro@czbiohub.org>
|
|
6
|
-
Maintainer-email: Talon Chandler <talon.chandler@czbiohub.org>, Shalin Mehta <shalin.mehta@czbiohub.org>
|
|
7
|
-
License: BSD 3-Clause License
|
|
8
|
-
|
|
9
|
-
Copyright (c) 2019, Chan Zuckerberg Biohub
|
|
10
|
-
|
|
11
|
-
Redistribution and use in source and binary forms, with or without
|
|
12
|
-
modification, are permitted provided that the following conditions are met:
|
|
13
|
-
|
|
14
|
-
1. Redistributions of source code must retain the above copyright notice, this
|
|
15
|
-
list of conditions and the following disclaimer.
|
|
16
|
-
|
|
17
|
-
2. Redistributions in binary form must reproduce the above copyright notice,
|
|
18
|
-
this list of conditions and the following disclaimer in the documentation
|
|
19
|
-
and/or other materials provided with the distribution.
|
|
20
|
-
|
|
21
|
-
3. Neither the name of the copyright holder nor the names of its
|
|
22
|
-
contributors may be used to endorse or promote products derived from
|
|
23
|
-
this software without specific prior written permission.
|
|
24
|
-
|
|
25
|
-
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
|
|
26
|
-
AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
|
|
27
|
-
IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
|
|
28
|
-
DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE
|
|
29
|
-
FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL
|
|
30
|
-
DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR
|
|
31
|
-
SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER
|
|
32
|
-
CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,
|
|
33
|
-
OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE
|
|
34
|
-
OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
|
|
35
|
-
|
|
36
|
-
Project-URL: Homepage, https://github.com/mehta-lab/waveorder
|
|
37
|
-
Project-URL: Repository, https://github.com/mehta-lab/waveorder
|
|
38
|
-
Project-URL: Issues, https://github.com/mehta-lab/waveorder/issues
|
|
39
|
-
Keywords: simulation,optics,phase,scattering,polarization,label-free,permittivity,reconstruction-algorithm,qlipp,mipolscope,permittivity-tensor-imaging
|
|
40
|
-
Classifier: Development Status :: 4 - Beta
|
|
41
|
-
Classifier: Intended Audience :: Science/Research
|
|
42
|
-
Classifier: License :: OSI Approved :: BSD License
|
|
43
|
-
Classifier: Programming Language :: Python :: 3
|
|
44
|
-
Classifier: Programming Language :: Python :: 3.10
|
|
45
|
-
Classifier: Programming Language :: Python :: 3.11
|
|
46
|
-
Classifier: Programming Language :: Python :: 3.12
|
|
47
|
-
Classifier: Topic :: Scientific/Engineering
|
|
48
|
-
Classifier: Topic :: Scientific/Engineering :: Image Processing
|
|
49
|
-
Classifier: Topic :: Scientific/Engineering :: Bio-Informatics
|
|
50
|
-
Classifier: Operating System :: POSIX :: Linux
|
|
51
|
-
Classifier: Operating System :: Microsoft :: Windows
|
|
52
|
-
Classifier: Operating System :: MacOS
|
|
53
|
-
Requires-Python: >=3.10
|
|
54
|
-
Description-Content-Type: text/markdown
|
|
55
|
-
License-File: LICENSE
|
|
56
|
-
Requires-Dist: numpy>=1.24
|
|
57
|
-
Requires-Dist: matplotlib>=3.1.1
|
|
58
|
-
Requires-Dist: scipy>=1.3.0
|
|
59
|
-
Requires-Dist: pywavelets>=1.1.1
|
|
60
|
-
Requires-Dist: ipywidgets>=7.5.1
|
|
61
|
-
Requires-Dist: torch>=2.4.1
|
|
62
|
-
Provides-Extra: dev
|
|
63
|
-
Requires-Dist: pytest; extra == "dev"
|
|
64
|
-
Requires-Dist: pytest-cov; extra == "dev"
|
|
65
|
-
Requires-Dist: black==25.1.0; extra == "dev"
|
|
66
|
-
Provides-Extra: examples
|
|
67
|
-
Requires-Dist: napari[all]; extra == "examples"
|
|
68
|
-
Requires-Dist: jupyter; extra == "examples"
|
|
69
|
-
Dynamic: license-file
|
|
70
|
-
|
|
71
|
-
# waveorder
|
|
72
|
-
|
|
73
|
-
[](https://pypi.org/project/waveorder)
|
|
74
|
-
[](https://pypistats.org/packages/waveorder)
|
|
75
|
-
[](https://pepy.tech/project/waveorder)
|
|
76
|
-
[](https://github.com/mehta-lab/waveorder/graphs/contributors)
|
|
77
|
-

|
|
78
|
-

|
|
79
|
-

|
|
80
|
-
|
|
81
|
-
|
|
82
|
-
This computational imaging library enables wave-optical simulation and reconstruction of optical properties that report microscopic architectural order.
|
|
83
|
-
|
|
84
|
-
## Computational label-agnostic imaging
|
|
85
|
-
|
|
86
|
-
https://github.com/user-attachments/assets/4f9969e5-94ce-4e08-9f30-68314a905db6
|
|
87
|
-
|
|
88
|
-
`waveorder` enables simulations and reconstructions of label-agnostic microscopy data as described in the following [preprint](https://arxiv.org/abs/2412.09775)
|
|
89
|
-
<details>
|
|
90
|
-
<summary> Chandler et al. 2024 </summary>
|
|
91
|
-
<pre><code>
|
|
92
|
-
@article{chandler_2024,
|
|
93
|
-
author = {Chandler, Talon and Hirata-Miyasaki, Eduardo and Ivanov, Ivan E. and Liu, Ziwen and Sundarraman, Deepika and Ryan, Allyson Quinn and Jacobo, Adrian and Balla, Keir and Mehta, Shalin B.},
|
|
94
|
-
title = {waveOrder: generalist framework for label-agnostic computational microscopy},
|
|
95
|
-
journal = {arXiv},
|
|
96
|
-
year = {2024},
|
|
97
|
-
month = dec,
|
|
98
|
-
eprint = {2412.09775},
|
|
99
|
-
doi = {10.48550/arXiv.2412.09775}
|
|
100
|
-
}
|
|
101
|
-
</code></pre>
|
|
102
|
-
</details>
|
|
103
|
-
|
|
104
|
-
Specifically, `waveorder` enables simulation and reconstruction of 2D or 3D:
|
|
105
|
-
|
|
106
|
-
1. __phase, projected retardance, and in-plane orientation__ from a polarization-diverse volumetric brightfield acquisition ([QLIPP](https://elifesciences.org/articles/55502)),
|
|
107
|
-
|
|
108
|
-
2. __phase__ from a volumetric brightfield acquisition ([2D phase](https://www.osapublishing.org/ao/abstract.cfm?uri=ao-54-28-8566)/[3D phase](https://www.osapublishing.org/ao/abstract.cfm?uri=ao-57-1-a205)),
|
|
109
|
-
|
|
110
|
-
3. __phase__ from an illumination-diverse volumetric acquisition ([2D](https://www.osapublishing.org/oe/fulltext.cfm?uri=oe-23-9-11394&id=315599)/[3D](https://www.osapublishing.org/boe/fulltext.cfm?uri=boe-7-10-3940&id=349951) differential phase contrast),
|
|
111
|
-
|
|
112
|
-
4. __fluorescence density__ from a widefield volumetric fluorescence acquisition (fluorescence deconvolution).
|
|
113
|
-
|
|
114
|
-
The [examples](https://github.com/mehta-lab/waveorder/tree/main/examples) demonstrate simulations and reconstruction for 2D QLIPP, 3D PODT, 3D fluorescence deconvolution, and 2D/3D PTI methods.
|
|
115
|
-
|
|
116
|
-
If you are interested in deploying QLIPP, phase from brightfield, or fluorescence deconvolution for label-agnostic imaging at scale, checkout our [napari plugin](https://www.napari-hub.org/plugins/recOrder-napari), [`recOrder-napari`](https://github.com/mehta-lab/recOrder).
|
|
117
|
-
|
|
118
|
-
## Permittivity tensor imaging
|
|
119
|
-
|
|
120
|
-
Additionally, `waveorder` enabled the development of a new label-free imaging method, __permittivity tensor imaging (PTI)__, that measures density and 3D orientation of biomolecules with diffraction-limited resolution. These measurements are reconstructed from polarization-resolved images acquired with a sequence of oblique illuminations.
|
|
121
|
-
|
|
122
|
-
The acquisition, calibration, background correction, reconstruction, and applications of PTI are described in the following [paper](https://doi.org/10.1101/2020.12.15.422951) published in Nature Methods:
|
|
123
|
-
|
|
124
|
-
<details>
|
|
125
|
-
<summary> Yeh et al. 2024 </summary>
|
|
126
|
-
<pre><code>
|
|
127
|
-
@article{yeh_2024,
|
|
128
|
-
author = {Yeh, Li-Hao and Ivanov, Ivan E. and Chandler, Talon and Byrum, Janie R. and Chhun, Bryant B. and Guo, Syuan-Ming and Foltz, Cameron and Hashemi, Ezzat and Perez-Bermejo, Juan A. and Wang, Huijun and Yu, Yanhao and Kazansky, Peter G. and Conklin, Bruce R. and Han, May H. and Mehta, Shalin B.},
|
|
129
|
-
title = {Permittivity tensor imaging: modular label-free imaging of 3D dry mass and 3D orientation at high resolution},
|
|
130
|
-
journal = {Nature Methods},
|
|
131
|
-
volume = {21},
|
|
132
|
-
number = {7},
|
|
133
|
-
pages = {1257--1274},
|
|
134
|
-
year = {2024},
|
|
135
|
-
month = jul,
|
|
136
|
-
issn = {1548-7105},
|
|
137
|
-
publisher = {Nature Publishing Group},
|
|
138
|
-
doi = {10.1038/s41592-024-02291-w}
|
|
139
|
-
}
|
|
140
|
-
</code></pre>
|
|
141
|
-
</details>
|
|
142
|
-
|
|
143
|
-
PTI provides volumetric reconstructions of mean permittivity ($\propto$ material density), differential permittivity ($\propto$ material anisotropy), 3D orientation, and optic sign. The following figure summarizes PTI acquisition and reconstruction with a small optical section of the mouse brain tissue:
|
|
144
|
-
|
|
145
|
-

|
|
146
|
-
|
|
147
|
-
## Examples
|
|
148
|
-
The [examples](https://github.com/mehta-lab/waveorder/tree/main/examples) illustrate simulations and reconstruction for 2D QLIPP, 3D phase from brightfield, and 2D/3D PTI methods.
|
|
149
|
-
|
|
150
|
-
If you are interested in deploying QLIPP or phase from brightbrield, or fluorescence deconvolution for label-agnostic imaging at scale, checkout our [napari plugin](https://www.napari-hub.org/plugins/recOrder-napari), [`recOrder-napari`](https://github.com/mehta-lab/recOrder).
|
|
151
|
-
|
|
152
|
-
## Citation
|
|
153
|
-
|
|
154
|
-
Please cite this repository, along with the relevant preprint or paper, if you use or adapt this code. The citation information can be found by clicking "Cite this repository" button in the About section in the right sidebar.
|
|
155
|
-
|
|
156
|
-
## Installation
|
|
157
|
-
|
|
158
|
-
Create a virtual environment:
|
|
159
|
-
|
|
160
|
-
```sh
|
|
161
|
-
conda create -y -n waveorder python=3.10
|
|
162
|
-
conda activate waveorder
|
|
163
|
-
```
|
|
164
|
-
|
|
165
|
-
Install `waveorder` from PyPI:
|
|
166
|
-
|
|
167
|
-
```sh
|
|
168
|
-
pip install waveorder
|
|
169
|
-
```
|
|
170
|
-
|
|
171
|
-
Use `waveorder` in your scripts:
|
|
172
|
-
|
|
173
|
-
```sh
|
|
174
|
-
python
|
|
175
|
-
>>> import waveorder
|
|
176
|
-
```
|
|
177
|
-
|
|
178
|
-
(Optional) Install example dependencies, clone the repository, and run an example script:
|
|
179
|
-
```sh
|
|
180
|
-
pip install waveorder[examples]
|
|
181
|
-
git clone https://github.com/mehta-lab/waveorder.git
|
|
182
|
-
python waveorder/examples/models/phase_thick_3d.py
|
|
183
|
-
```
|
|
184
|
-
|
|
185
|
-
(M1 users) `pytorch` has [incomplete GPU support](https://github.com/pytorch/pytorch/issues/77764),
|
|
186
|
-
so please use `export PYTORCH_ENABLE_MPS_FALLBACK=1`
|
|
187
|
-
to allow some operators to fallback to CPU
|
|
188
|
-
if you plan to use GPU acceleration for polarization reconstruction.
|