pylocuszoom 1.2.0__py3-none-any.whl → 1.3.1__py3-none-any.whl
This diff represents the content of publicly available package versions that have been released to one of the supported registries. The information contained in this diff is provided for informational purposes only and reflects changes between package versions as they appear in their respective public registries.
- pylocuszoom/__init__.py +16 -2
- pylocuszoom/backends/base.py +94 -2
- pylocuszoom/backends/bokeh_backend.py +160 -6
- pylocuszoom/backends/matplotlib_backend.py +142 -2
- pylocuszoom/backends/plotly_backend.py +101 -1
- pylocuszoom/coloc.py +82 -0
- pylocuszoom/coloc_plotter.py +390 -0
- pylocuszoom/colors.py +26 -0
- pylocuszoom/config.py +61 -0
- pylocuszoom/labels.py +41 -16
- pylocuszoom/ld.py +239 -0
- pylocuszoom/ld_heatmap_plotter.py +252 -0
- pylocuszoom/miami_plotter.py +490 -0
- pylocuszoom/plotter.py +472 -6
- {pylocuszoom-1.2.0.dist-info → pylocuszoom-1.3.1.dist-info}/METADATA +166 -21
- {pylocuszoom-1.2.0.dist-info → pylocuszoom-1.3.1.dist-info}/RECORD +18 -14
- pylocuszoom-1.3.1.dist-info/licenses/LICENSE.md +595 -0
- pylocuszoom-1.2.0.dist-info/licenses/LICENSE.md +0 -17
- {pylocuszoom-1.2.0.dist-info → pylocuszoom-1.3.1.dist-info}/WHEEL +0 -0
|
@@ -1,6 +1,6 @@
|
|
|
1
1
|
Metadata-Version: 2.4
|
|
2
2
|
Name: pylocuszoom
|
|
3
|
-
Version: 1.
|
|
3
|
+
Version: 1.3.1
|
|
4
4
|
Summary: Publication-ready regional association plots with LD coloring, gene tracks, and recombination overlays
|
|
5
5
|
Project-URL: Homepage, https://github.com/michael-denyer/pylocuszoom
|
|
6
6
|
Project-URL: Documentation, https://github.com/michael-denyer/pylocuszoom#readme
|
|
@@ -72,20 +72,23 @@ Inspired by [LocusZoom](http://locuszoom.org/) and [locuszoomr](https://github.c
|
|
|
72
72
|
- **SNP labels (matplotlib)**: Automatic labeling of top SNPs by p-value (RS IDs)
|
|
73
73
|
- **Hover tooltips (Plotly and Bokeh)**: Detailed SNP data on hover
|
|
74
74
|
|
|
75
|
-

|
|
75
|
+
|
|
76
76
|
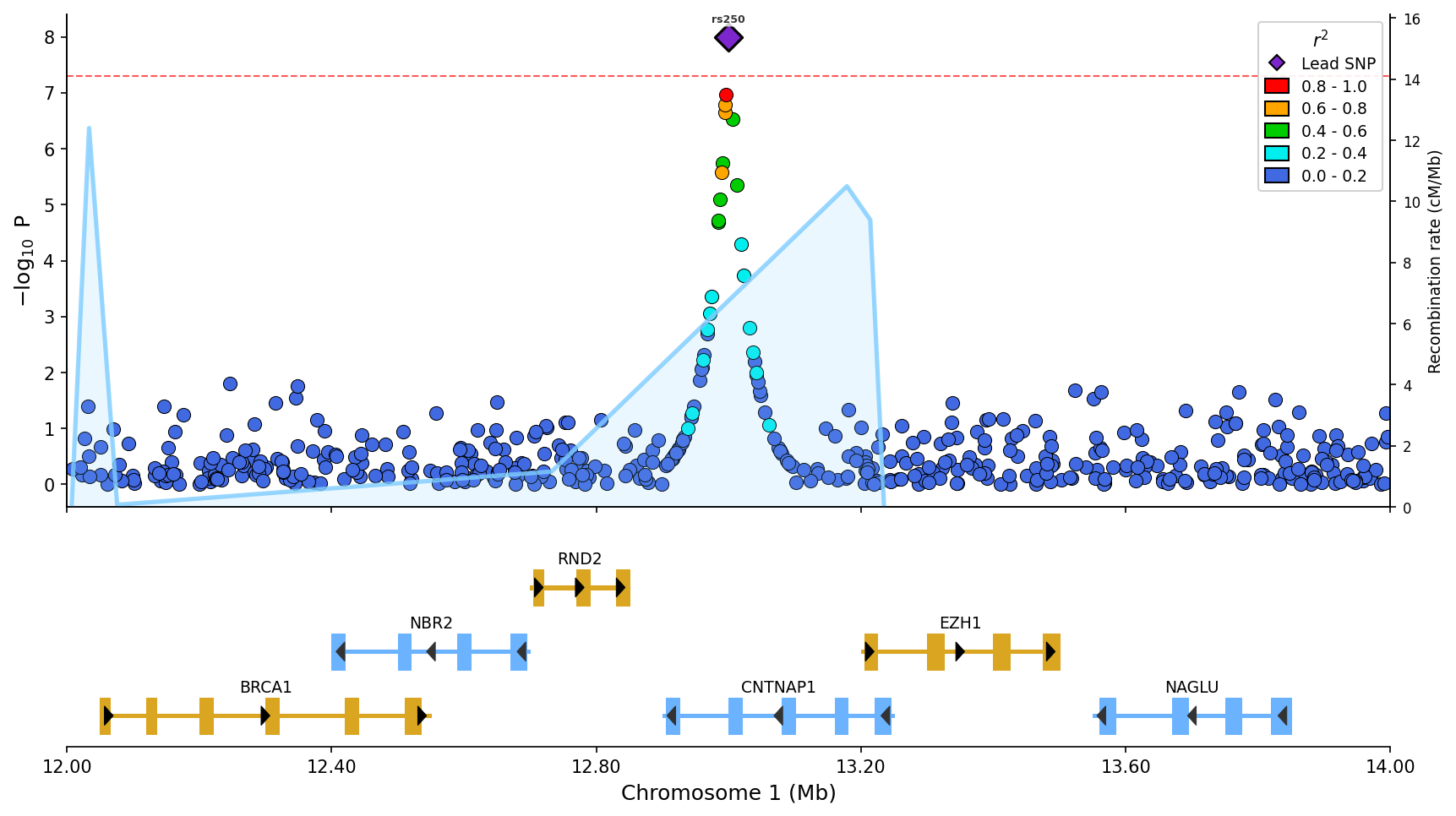
*Regional association plot with LD coloring, gene/exon track, recombination rate overlay (blue line), and top SNP labels.*
|
|
77
77
|
|
|
78
78
|
2. **Stacked plots**: Compare multiple GWAS/phenotypes vertically
|
|
79
|
-
3. **
|
|
80
|
-
4. **
|
|
81
|
-
5. **
|
|
82
|
-
6. **
|
|
83
|
-
7. **
|
|
84
|
-
8. **
|
|
85
|
-
9. **
|
|
86
|
-
10. **
|
|
87
|
-
11. **
|
|
88
|
-
12. **
|
|
79
|
+
3. **Miami plots**: Mirrored Manhattan plots for comparing two GWAS datasets (discovery vs replication)
|
|
80
|
+
4. **Manhattan plots**: Genome-wide association visualization with chromosome coloring
|
|
81
|
+
5. **QQ plots**: Quantile-quantile plots with confidence bands and genomic inflation factor
|
|
82
|
+
6. **eQTL plot**: Expression QTL data aligned with association plots and gene tracks
|
|
83
|
+
7. **Fine-mapping plots**: Visualize SuSiE credible sets with posterior inclusion probabilities
|
|
84
|
+
8. **PheWAS plots**: Phenome-wide association study visualization across multiple phenotypes
|
|
85
|
+
9. **Forest plots**: Meta-analysis effect size visualization with confidence intervals
|
|
86
|
+
10. **LD heatmaps**: Triangular heatmaps showing pairwise LD patterns, standalone or integrated below regional plots
|
|
87
|
+
11. **Colocalization plots**: GWAS-eQTL scatter plots with LD coloring, correlation statistics, and effect direction visualization
|
|
88
|
+
12. **Multiple backends**: matplotlib (publication-ready), plotly (interactive), bokeh (dashboard integration)
|
|
89
|
+
12. **Pandas and PySpark support**: Works with both Pandas and PySpark DataFrames for large-scale genomics data
|
|
90
|
+
13. **Convenience data file loaders**: Load and validate common GWAS, eQTL and fine-mapping file formats
|
|
91
|
+
14. **Automatic gene annotations**: Fetch gene/exon data from Ensembl REST API with caching (human, mouse, rat, canine, feline, and any Ensembl species)
|
|
89
92
|
|
|
90
93
|
## Installation
|
|
91
94
|
|
|
@@ -255,7 +258,7 @@ fig = plotter.plot_stacked(
|
|
|
255
258
|
)
|
|
256
259
|
```
|
|
257
260
|
|
|
258
|
-

|
|
261
|
+
|
|
259
262
|
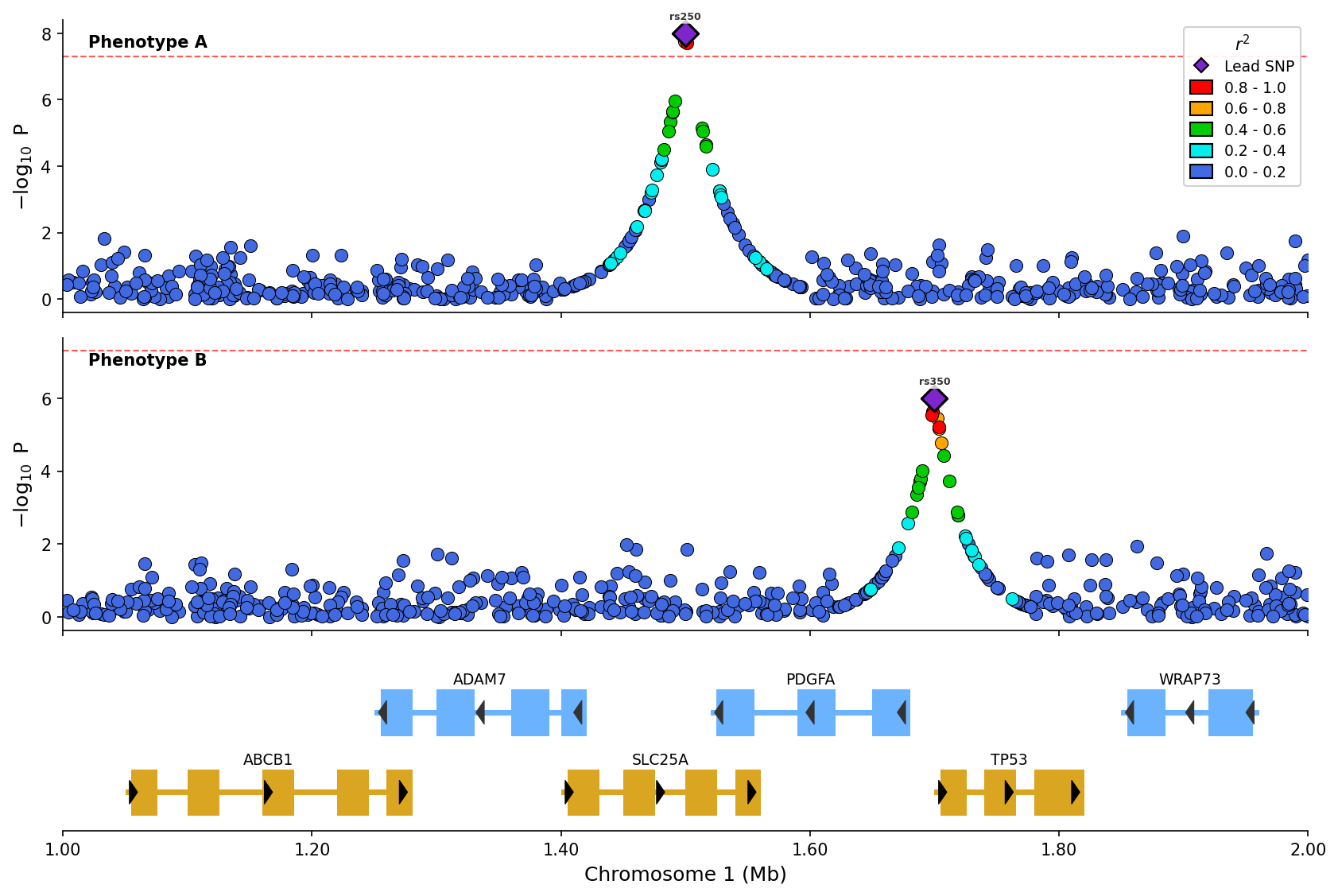
*Stacked plot comparing two phenotypes with LD coloring and shared gene track.*
|
|
260
263
|
|
|
261
264
|
## eQTL Overlay
|
|
@@ -284,7 +287,7 @@ fig = plotter.plot_stacked(
|
|
|
284
287
|
)
|
|
285
288
|
```
|
|
286
289
|
|
|
287
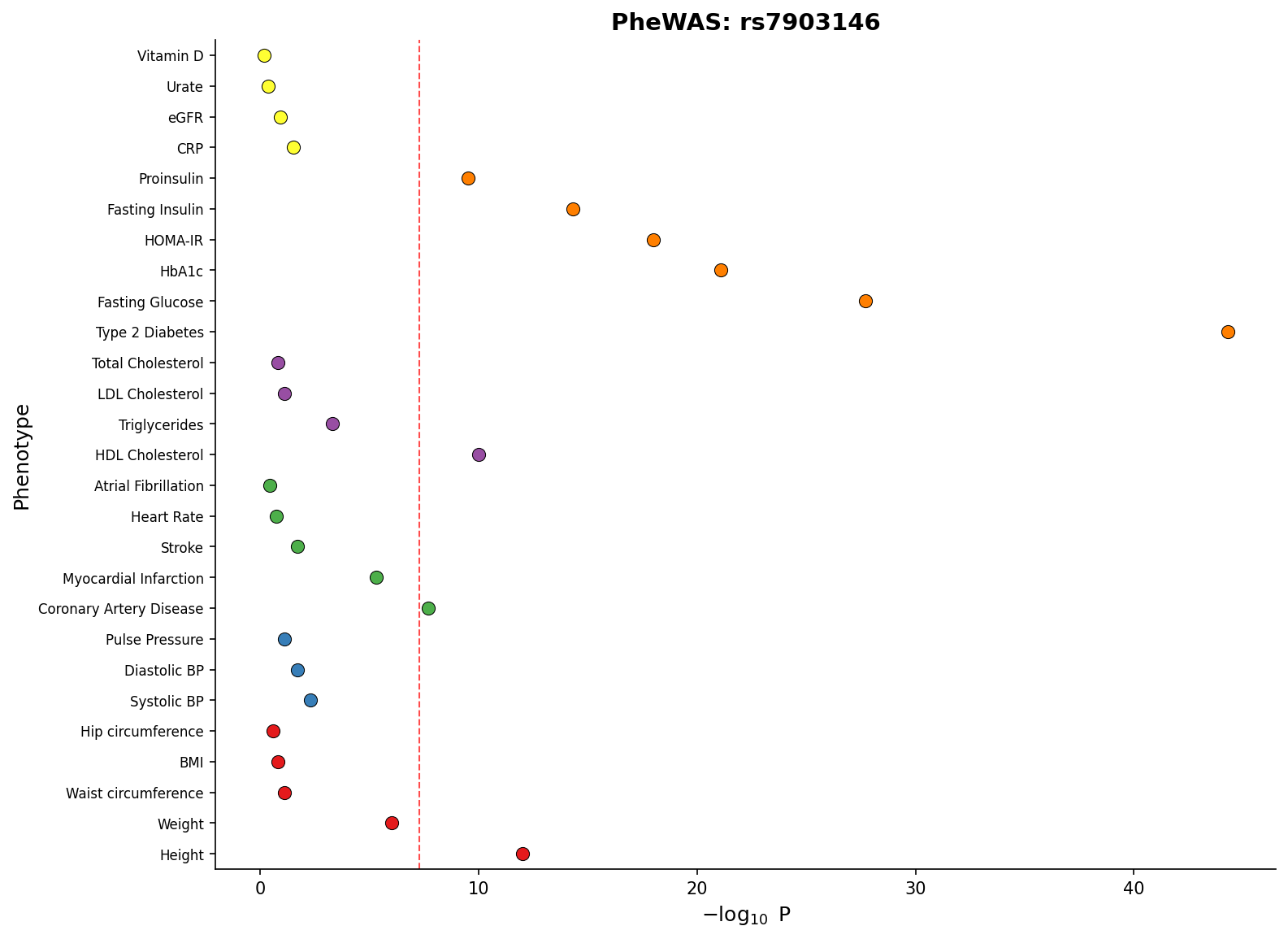
|
-

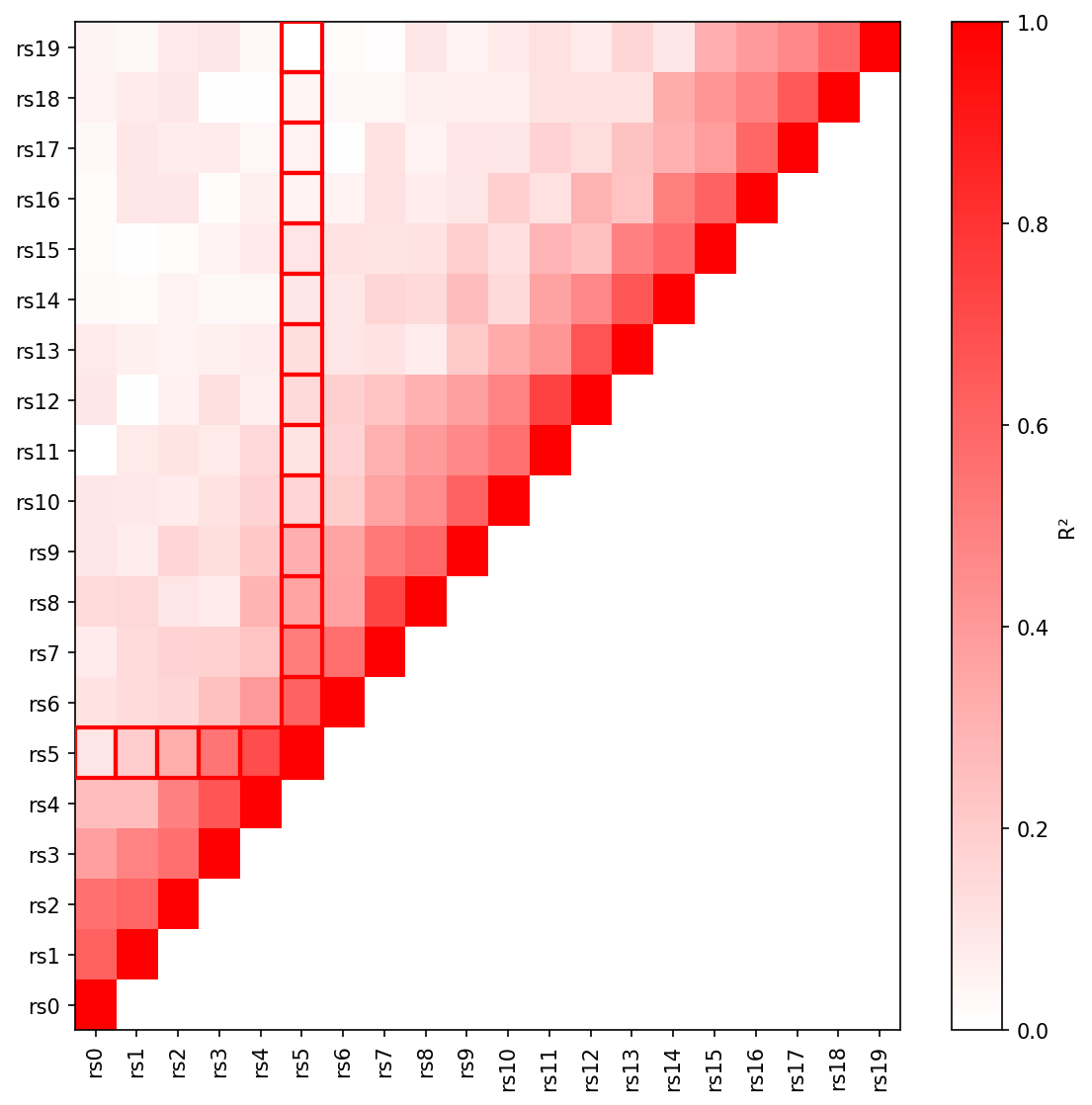
|
|
290
|
+
|
|
288
291
|
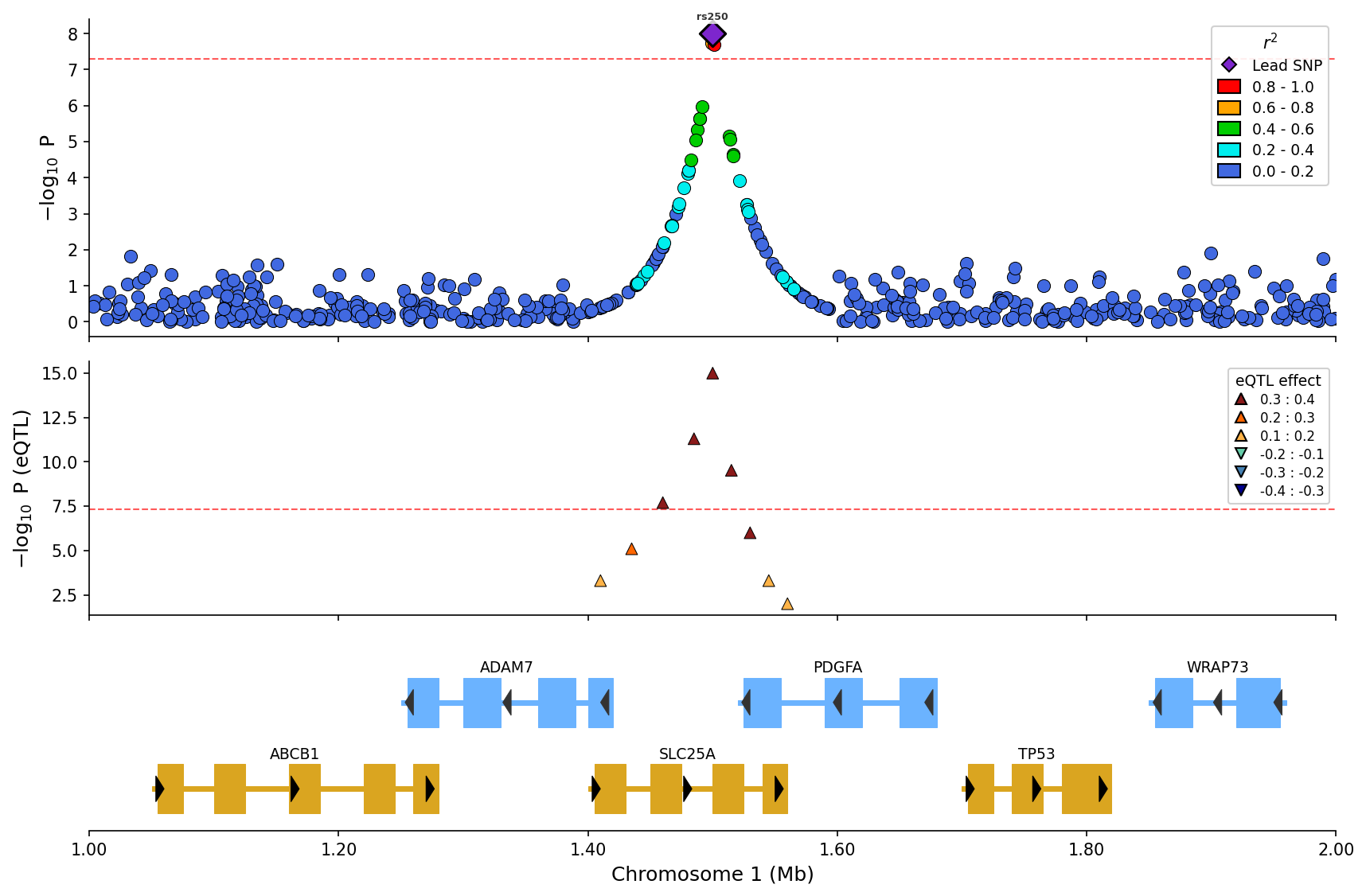
*eQTL overlay with effect direction (up/down triangles) and magnitude binning.*
|
|
289
292
|
|
|
290
293
|
## Fine-mapping Visualization
|
|
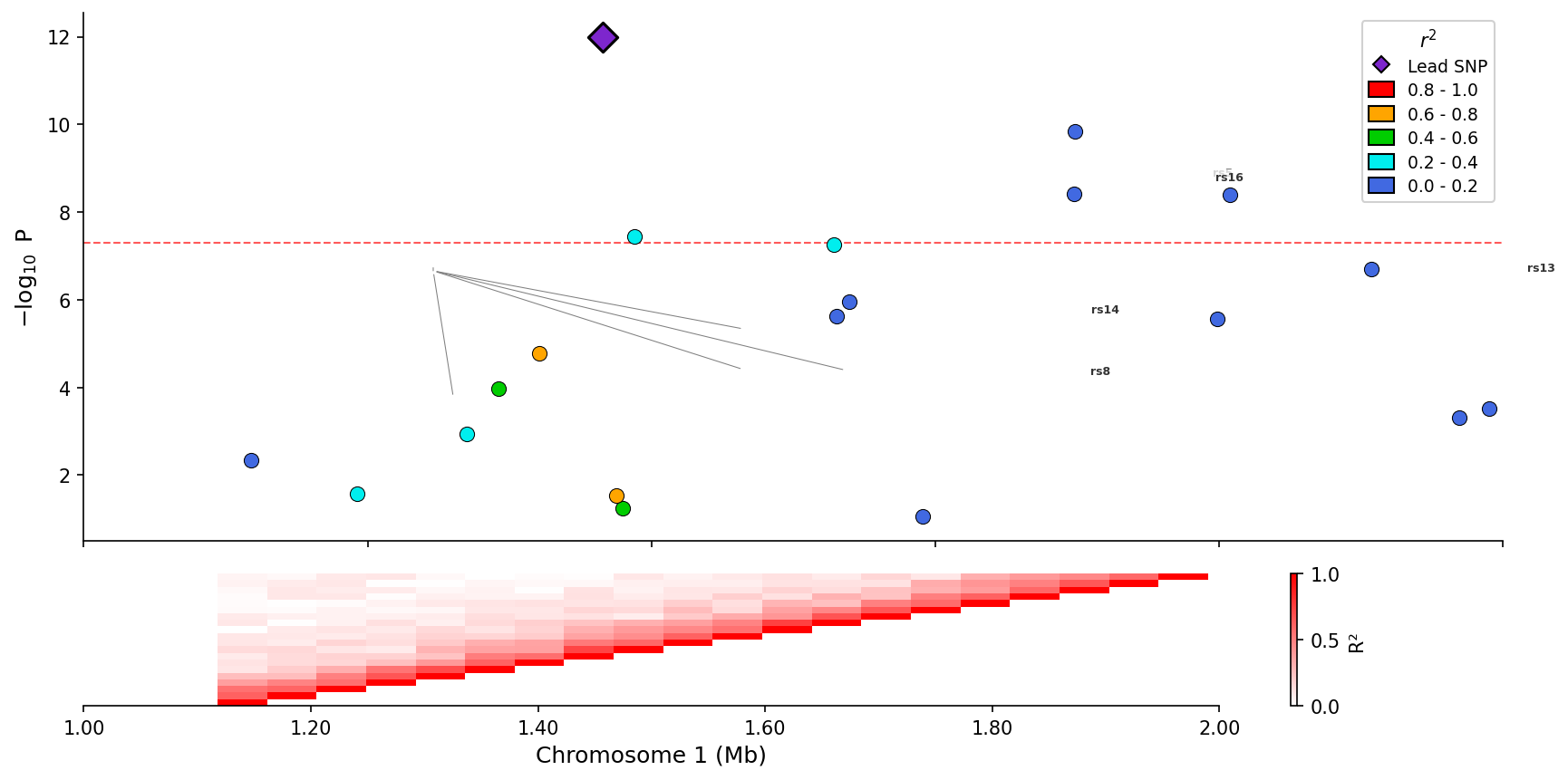
@@ -313,9 +316,108 @@ fig = plotter.plot_stacked(
|
|
|
313
316
|
)
|
|
314
317
|
```
|
|
315
318
|
|
|
316
|
-

|
|
319
|
+
|
|
317
320
|
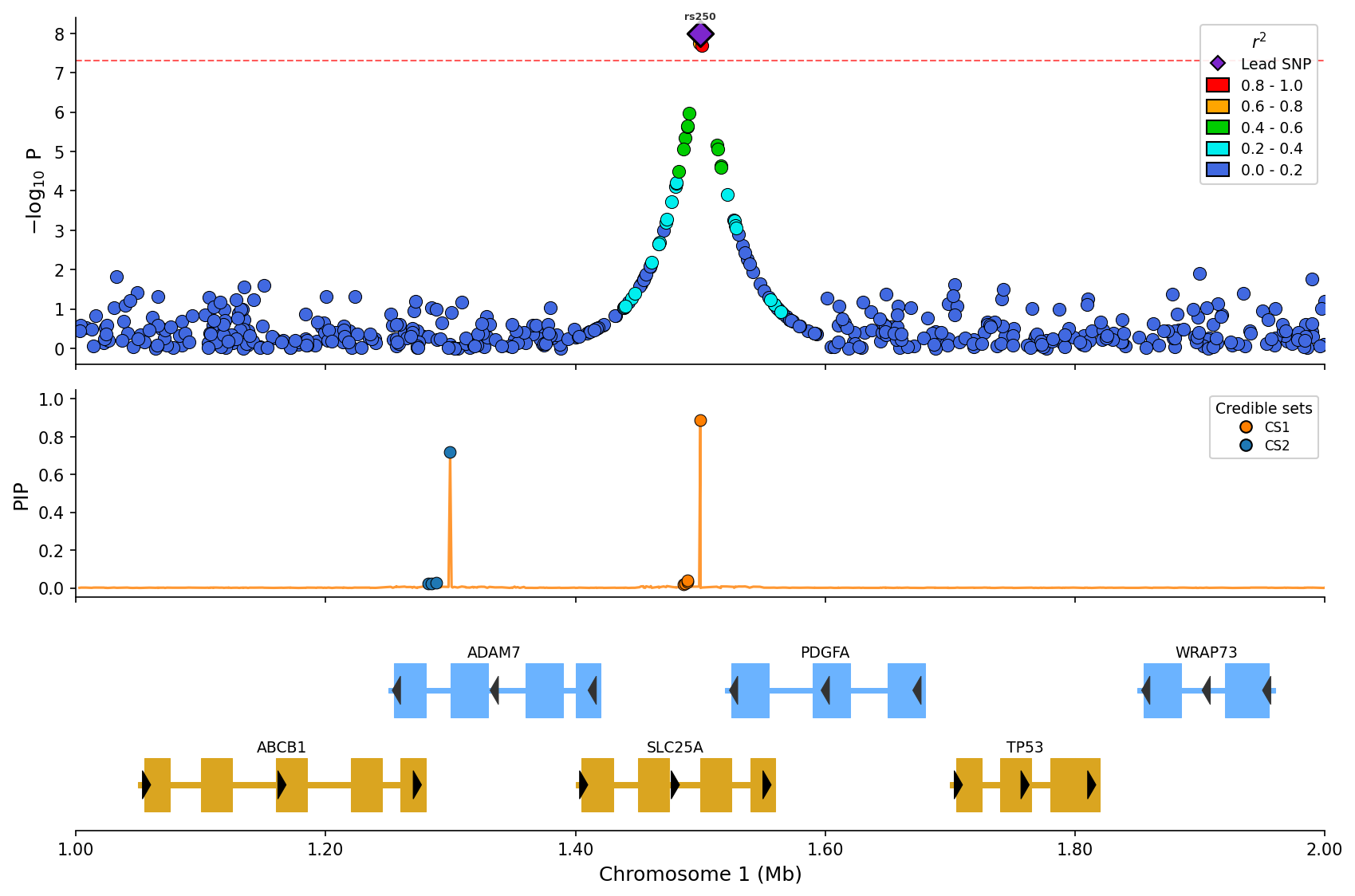
*Fine-mapping visualization with PIP line and credible set coloring (CS1/CS2).*
|
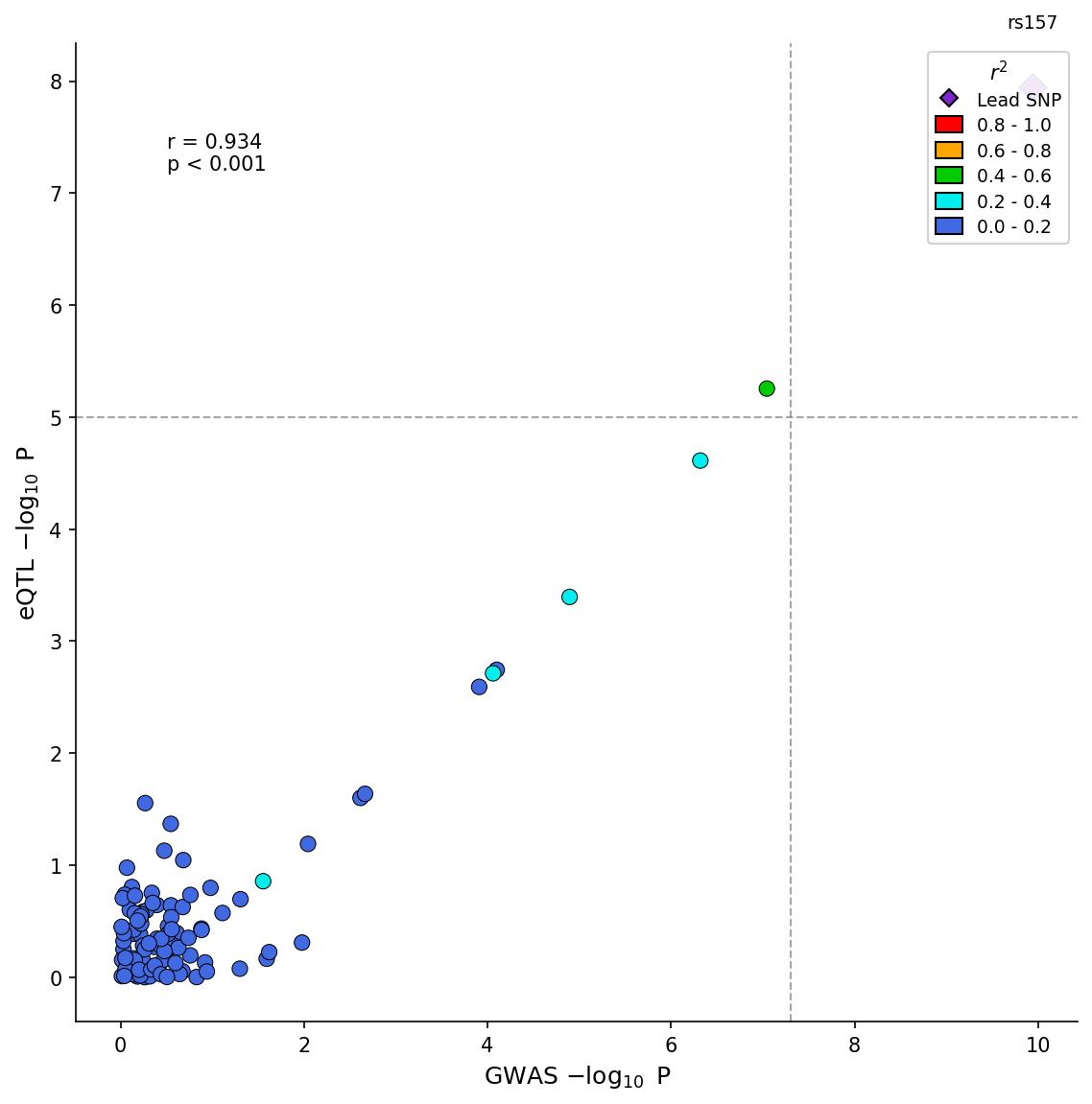
|
318
321
|
|
|
322
|
+
## LD Heatmaps
|
|
323
|
+
|
|
324
|
+
Create triangular LD heatmaps showing pairwise linkage disequilibrium patterns:
|
|
325
|
+
|
|
326
|
+
```python
|
|
327
|
+
from pylocuszoom import LDHeatmapPlotter
|
|
328
|
+
|
|
329
|
+
# ld_matrix is a square DataFrame with SNP IDs as index/columns
|
|
330
|
+
# snp_ids is a list of SNP IDs in the matrix
|
|
331
|
+
|
|
332
|
+
ld_plotter = LDHeatmapPlotter()
|
|
333
|
+
fig = ld_plotter.plot(
|
|
334
|
+
ld_matrix,
|
|
335
|
+
snp_ids,
|
|
336
|
+
highlight_snp_id="rs12345", # Highlight lead SNP
|
|
337
|
+
metric="r2", # or "dprime"
|
|
338
|
+
)
|
|
339
|
+
fig.savefig("ld_heatmap.png", dpi=150)
|
|
340
|
+
```
|
|
341
|
+
|
|
342
|
+
|
|
343
|
+
*Triangular LD heatmap with R² values and lead SNP highlighted.*
|
|
344
|
+
|
|
345
|
+
### Integrated LD Heatmap with Regional Plot
|
|
346
|
+
|
|
347
|
+
Add an LD heatmap panel below a regional association plot:
|
|
348
|
+
|
|
349
|
+
```python
|
|
350
|
+
from pylocuszoom import LocusZoomPlotter
|
|
351
|
+
|
|
352
|
+
plotter = LocusZoomPlotter(species="canine")
|
|
353
|
+
|
|
354
|
+
fig = plotter.plot(
|
|
355
|
+
gwas_df,
|
|
356
|
+
chrom=1,
|
|
357
|
+
start=1000000,
|
|
358
|
+
end=2000000,
|
|
359
|
+
lead_pos=1500000,
|
|
360
|
+
ld_heatmap_df=ld_matrix, # Pairwise LD matrix
|
|
361
|
+
ld_heatmap_snp_ids=snp_ids, # SNP IDs in matrix
|
|
362
|
+
ld_heatmap_height=0.25, # Panel height ratio
|
|
363
|
+
)
|
|
364
|
+
```
|
|
365
|
+
|
|
366
|
+
|
|
367
|
+
*Regional association plot with integrated LD heatmap panel below.*
|
|
368
|
+
|
|
369
|
+
## Colocalization Plots
|
|
370
|
+
|
|
371
|
+
Visualize GWAS-eQTL colocalization by comparing association signals in a scatter plot with LD coloring:
|
|
372
|
+
|
|
373
|
+
```python
|
|
374
|
+
from pylocuszoom import ColocPlotter
|
|
375
|
+
|
|
376
|
+
# GWAS and eQTL data with matching positions
|
|
377
|
+
gwas_df = pd.DataFrame({
|
|
378
|
+
"pos": positions,
|
|
379
|
+
"p": gwas_pvalues,
|
|
380
|
+
"ld_r2": ld_values, # Optional: LD with lead SNP
|
|
381
|
+
})
|
|
382
|
+
|
|
383
|
+
eqtl_df = pd.DataFrame({
|
|
384
|
+
"pos": positions,
|
|
385
|
+
"p": eqtl_pvalues,
|
|
386
|
+
})
|
|
387
|
+
|
|
388
|
+
plotter = ColocPlotter()
|
|
389
|
+
fig = plotter.plot_coloc(
|
|
390
|
+
gwas_df=gwas_df,
|
|
391
|
+
eqtl_df=eqtl_df,
|
|
392
|
+
pos_col="pos",
|
|
393
|
+
gwas_p_col="p",
|
|
394
|
+
eqtl_p_col="p",
|
|
395
|
+
ld_col="ld_r2",
|
|
396
|
+
gwas_threshold=5e-8,
|
|
397
|
+
eqtl_threshold=1e-5,
|
|
398
|
+
)
|
|
399
|
+
fig.savefig("colocalization.png", dpi=150)
|
|
400
|
+
```
|
|
401
|
+
|
|
402
|
+
|
|
403
|
+
*GWAS-eQTL colocalization scatter plot with LD coloring and correlation statistics.*
|
|
404
|
+
|
|
405
|
+
**Advanced options** include effect direction coloring and H4 posterior probability display:
|
|
406
|
+
|
|
407
|
+
```python
|
|
408
|
+
fig = plotter.plot_coloc(
|
|
409
|
+
gwas_df=gwas_df,
|
|
410
|
+
eqtl_df=eqtl_df,
|
|
411
|
+
pos_col="pos",
|
|
412
|
+
gwas_p_col="p",
|
|
413
|
+
eqtl_p_col="p",
|
|
414
|
+
gwas_effect_col="beta",
|
|
415
|
+
eqtl_effect_col="slope",
|
|
416
|
+
color_by_effect=True, # Green=congruent, Red=incongruent
|
|
417
|
+
h4_posterior=0.85, # Display coloc H4 probability
|
|
418
|
+
)
|
|
419
|
+
```
|
|
420
|
+
|
|
319
421
|
## PheWAS Plots
|
|
320
422
|
|
|
321
423
|
Visualize associations of a single variant across multiple phenotypes:
|
|
@@ -337,7 +439,7 @@ fig = stats_plotter.plot_phewas(
|
|
|
337
439
|
)
|
|
338
440
|
```
|
|
339
441
|
|
|
340
|
-

|
|
442
|
+
|
|
341
443
|
*PheWAS plot showing associations across phenotype categories with significance threshold.*
|
|
342
444
|
|
|
343
445
|
## Forest Plots
|
|
@@ -363,9 +465,52 @@ fig = stats_plotter.plot_forest(
|
|
|
363
465
|
)
|
|
364
466
|
```
|
|
365
467
|
|
|
366
|
-

|
|
468
|
+
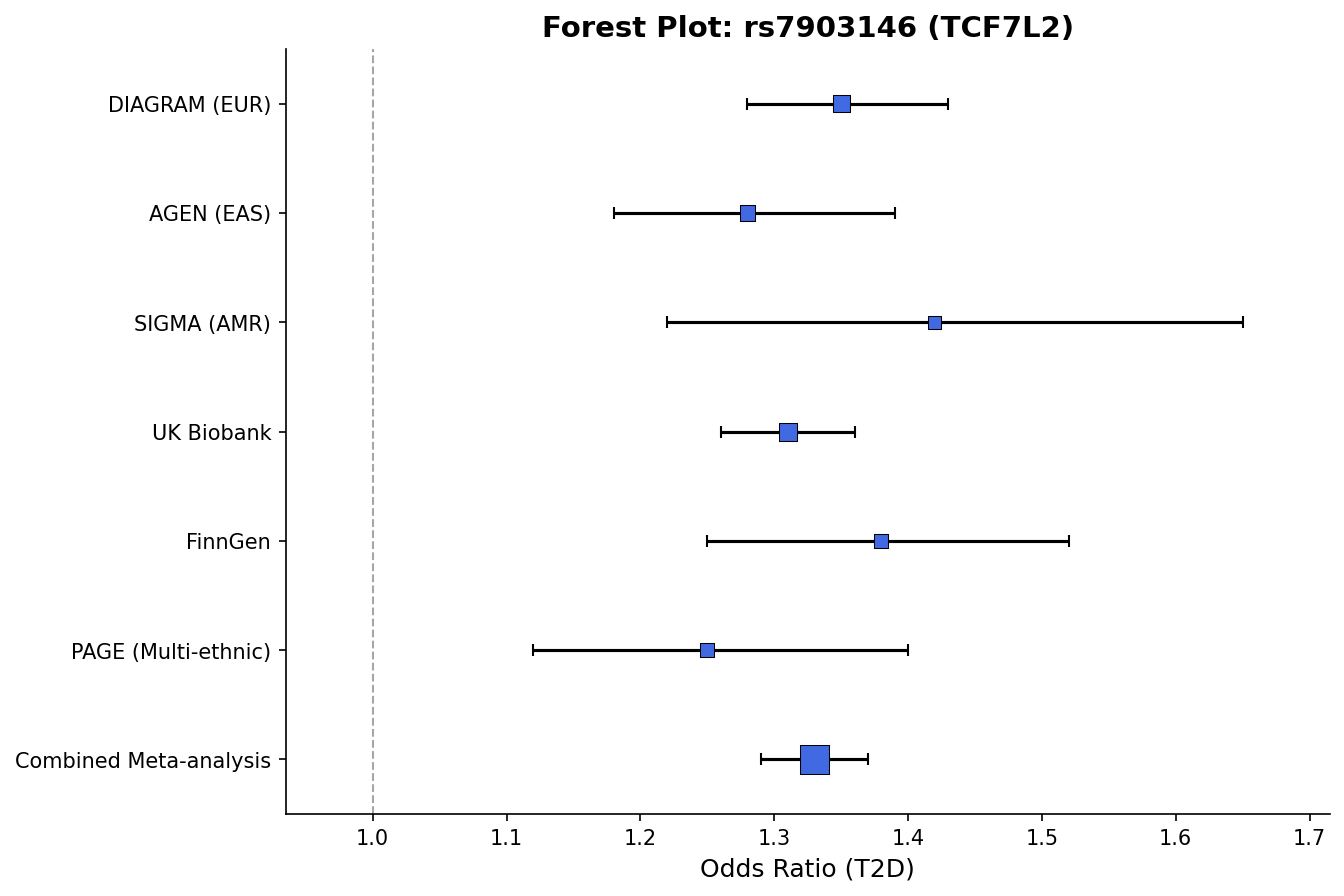
|
|
367
469
|
*Forest plot with effect sizes, confidence intervals, and weight-proportional markers.*
|
|
368
470
|
|
|
471
|
+
## Miami Plots
|
|
472
|
+
|
|
473
|
+
Compare two GWAS datasets with mirrored Manhattan plots (top panel ascending, bottom panel inverted):
|
|
474
|
+
|
|
475
|
+
```python
|
|
476
|
+
from pylocuszoom import MiamiPlotter
|
|
477
|
+
|
|
478
|
+
plotter = MiamiPlotter(species="human")
|
|
479
|
+
|
|
480
|
+
fig = plotter.plot_miami(
|
|
481
|
+
discovery_df,
|
|
482
|
+
replication_df,
|
|
483
|
+
chrom_col="chrom",
|
|
484
|
+
pos_col="pos",
|
|
485
|
+
p_col="p",
|
|
486
|
+
top_label="Discovery",
|
|
487
|
+
bottom_label="Replication",
|
|
488
|
+
top_threshold=5e-8,
|
|
489
|
+
bottom_threshold=1e-6,
|
|
490
|
+
highlight_regions=[("6", 30_000_000, 35_000_000)], # Highlight MHC region
|
|
491
|
+
)
|
|
492
|
+
fig.savefig("miami.png", dpi=150)
|
|
493
|
+
```
|
|
494
|
+
|
|
495
|
+
**Interactive backends** (Plotly/Bokeh) provide hover tooltips showing SNP details:
|
|
496
|
+
|
|
497
|
+
```python
|
|
498
|
+
# Plotly - interactive HTML with hover tooltips
|
|
499
|
+
plotter = MiamiPlotter(species="human", backend="plotly")
|
|
500
|
+
fig = plotter.plot_miami(discovery_df, replication_df, ...)
|
|
501
|
+
fig.write_html("miami_interactive.html")
|
|
502
|
+
|
|
503
|
+
# Bokeh - dashboard-ready interactive plots
|
|
504
|
+
from bokeh.io import output_file, save
|
|
505
|
+
plotter = MiamiPlotter(species="human", backend="bokeh")
|
|
506
|
+
fig = plotter.plot_miami(discovery_df, replication_df, ...)
|
|
507
|
+
output_file("miami_bokeh.html")
|
|
508
|
+
save(fig)
|
|
509
|
+
```
|
|
510
|
+
|
|
511
|
+
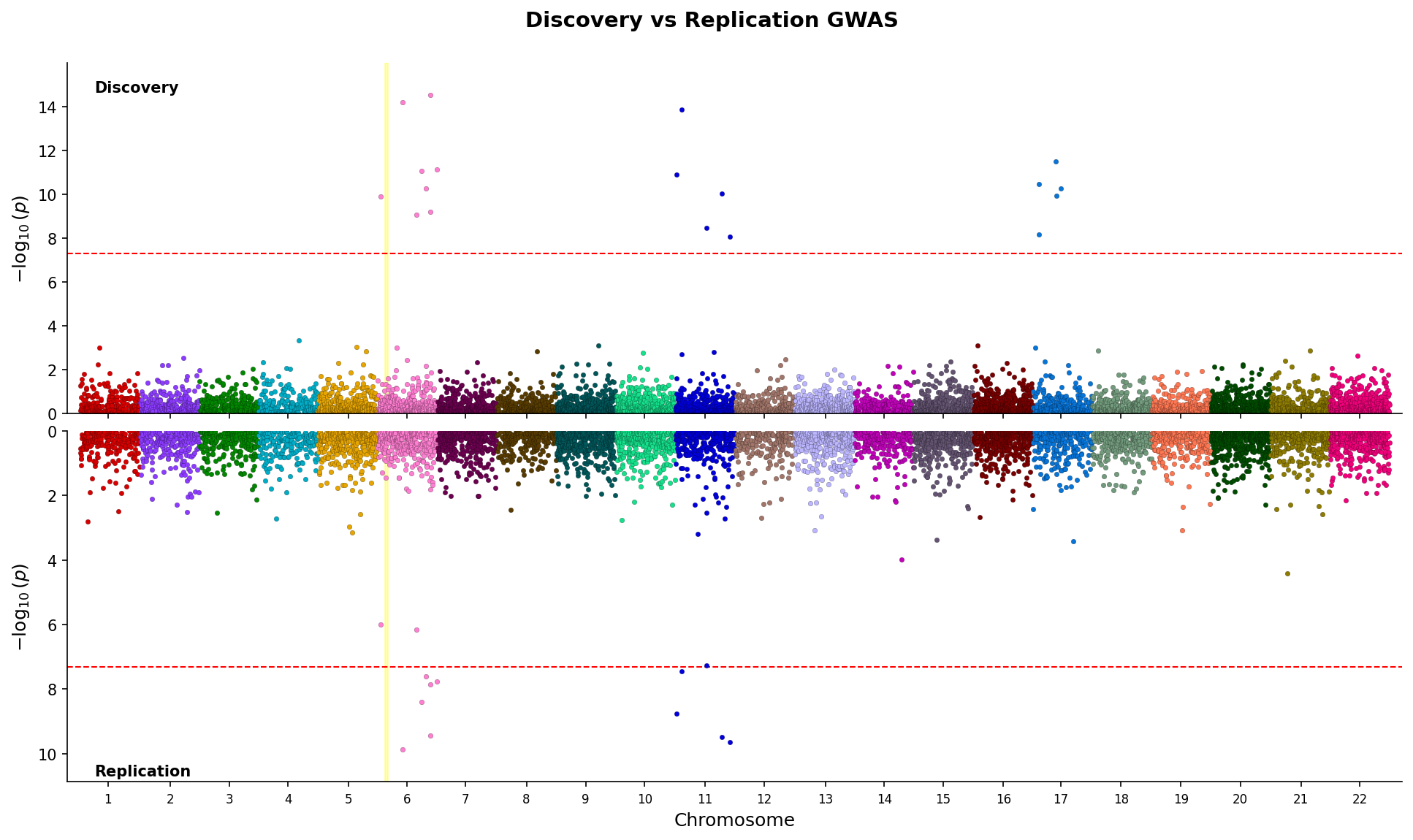
|
|
512
|
+
*Miami plot comparing discovery and replication GWAS with mirrored y-axes and region highlighting.*
|
|
513
|
+
|
|
369
514
|
## Manhattan Plots
|
|
370
515
|
|
|
371
516
|
Create genome-wide Manhattan plots showing associations across all chromosomes:
|
|
@@ -386,7 +531,7 @@ fig = plotter.plot_manhattan(
|
|
|
386
531
|
fig.savefig("manhattan.png", dpi=150)
|
|
387
532
|
```
|
|
388
533
|
|
|
389
|
-

|
|
534
|
+
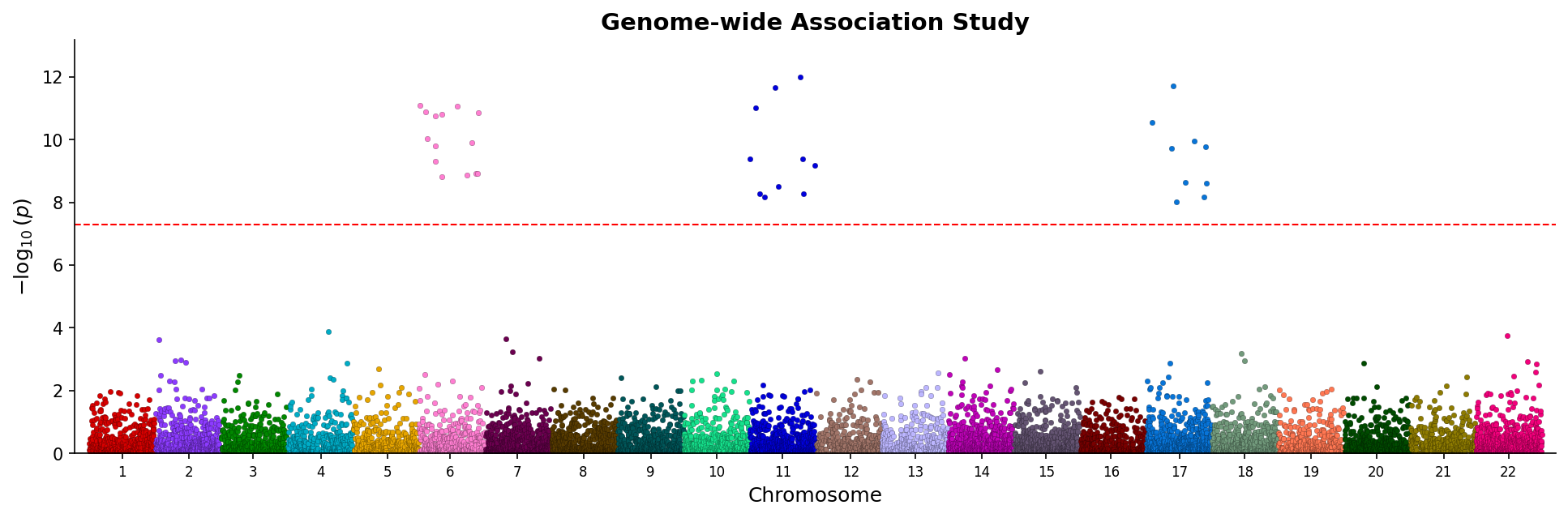
|
|
390
535
|
*Manhattan plot showing genome-wide associations with chromosome coloring and significance threshold.*
|
|
391
536
|
|
|
392
537
|
Categorical Manhattan plots (PheWAS-style) are also supported:
|
|
@@ -418,7 +563,7 @@ fig = plotter.plot_qq(
|
|
|
418
563
|
fig.savefig("qq_plot.png", dpi=150)
|
|
419
564
|
```
|
|
420
565
|
|
|
421
|
-

|
|
566
|
+
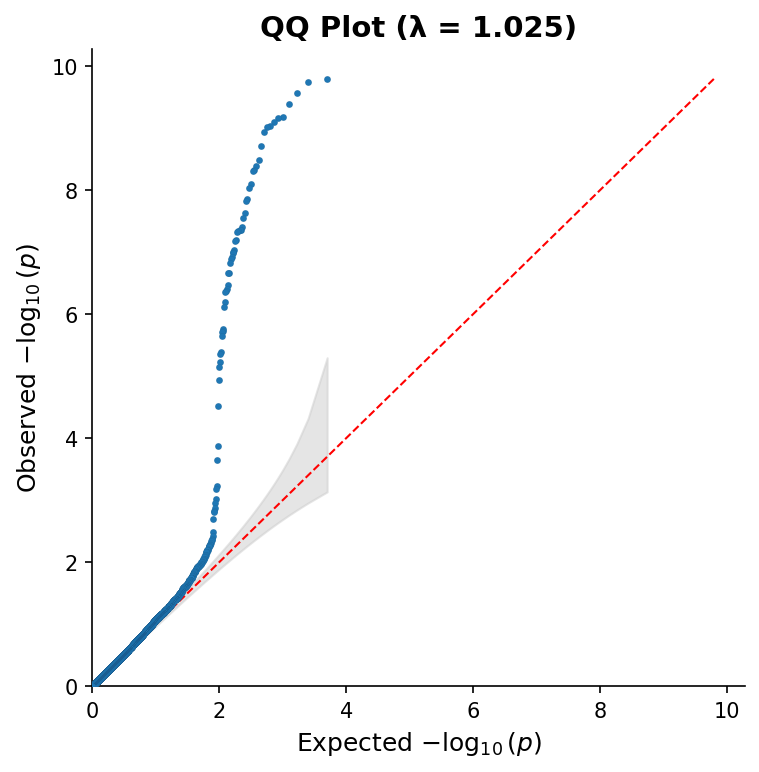
|
|
422
567
|
*QQ plot with 95% confidence band and genomic inflation factor (λ).*
|
|
423
568
|
|
|
424
569
|
## Stacked Manhattan Plots
|
|
@@ -443,7 +588,7 @@ fig = plotter.plot_manhattan_stacked(
|
|
|
443
588
|
fig.savefig("manhattan_stacked.png", dpi=150)
|
|
444
589
|
```
|
|
445
590
|
|
|
446
|
-

|
|
591
|
+
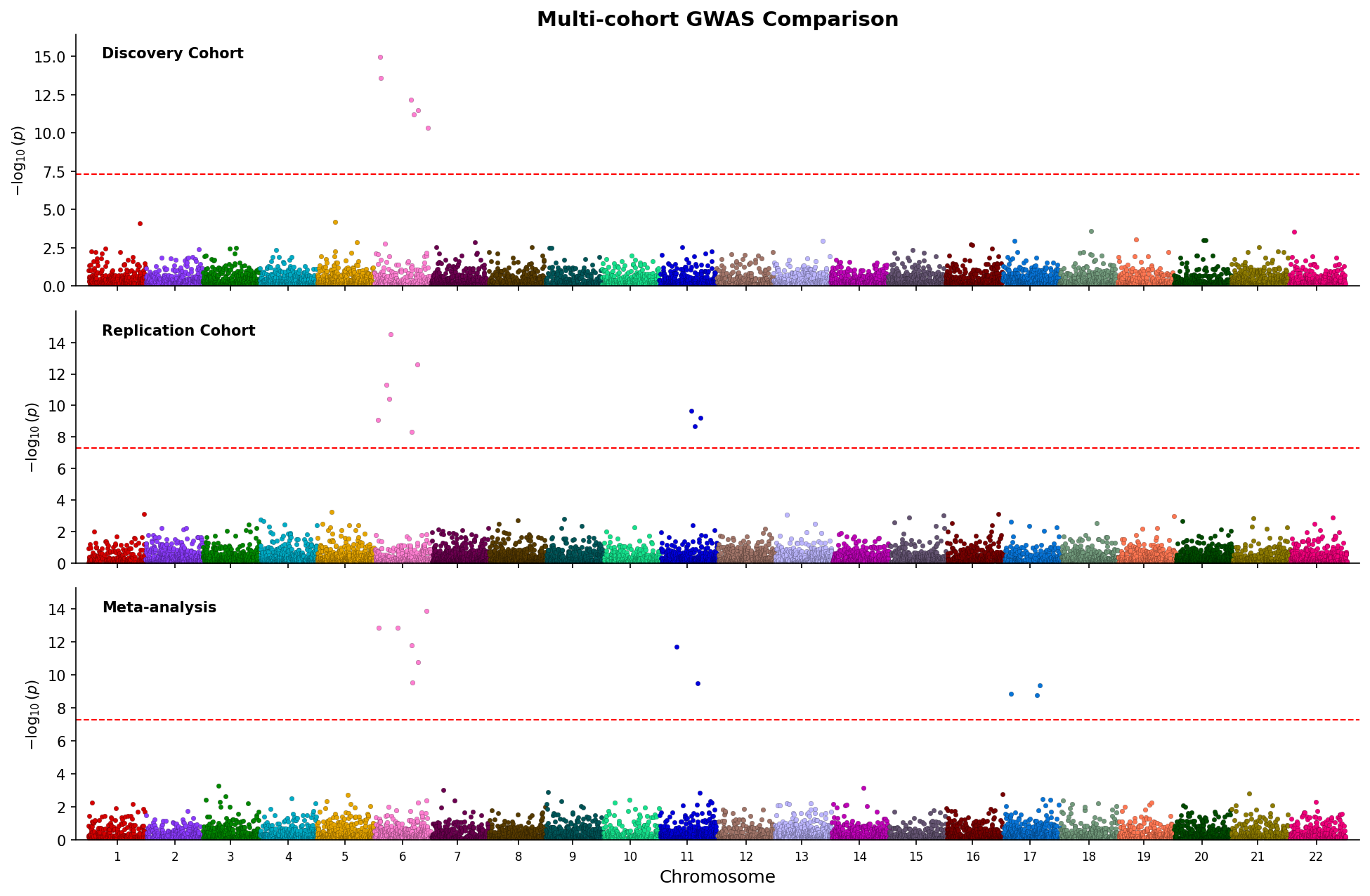
|
|
447
592
|
*Stacked Manhattan plots comparing three GWAS studies with shared chromosome axis.*
|
|
448
593
|
|
|
449
594
|
## Manhattan and QQ Side-by-Side
|
|
@@ -469,7 +614,7 @@ fig = plotter.plot_manhattan_qq(
|
|
|
469
614
|
fig.savefig("manhattan_qq.png", dpi=150)
|
|
470
615
|
```
|
|
471
616
|
|
|
472
|
-

|
|
617
|
+
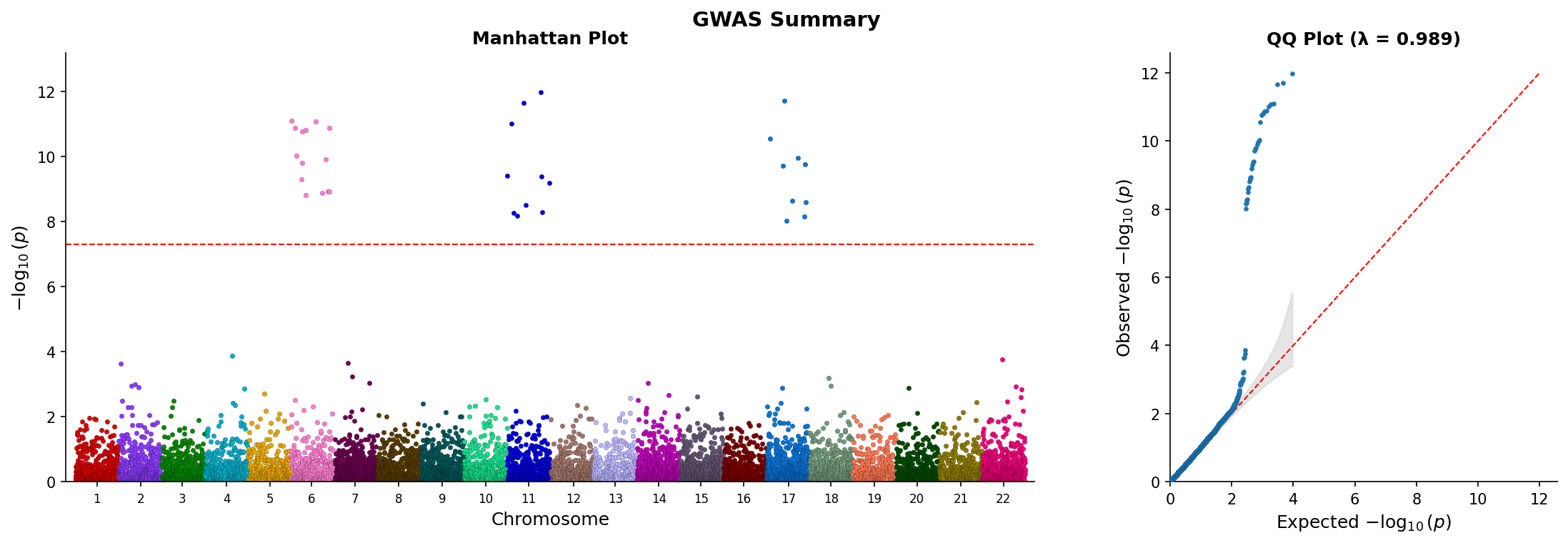
|
|
473
618
|
*Combined Manhattan and QQ plot showing genome-wide associations and p-value distribution.*
|
|
474
619
|
|
|
475
620
|
## PySpark Support
|
|
@@ -1,21 +1,25 @@
|
|
|
1
|
-
pylocuszoom/__init__.py,sha256=
|
|
1
|
+
pylocuszoom/__init__.py,sha256=VwZBIjjY-k6oQVkSgcsAwTkDitHdaHRe2gDUZ5b1YhA,6599
|
|
2
2
|
pylocuszoom/_plotter_utils.py,sha256=ELdSOcKk2KvOo_AxEWHeutmmUS4zZMaDMmQfpQUWaF0,1541
|
|
3
|
-
pylocuszoom/
|
|
4
|
-
pylocuszoom/
|
|
3
|
+
pylocuszoom/coloc.py,sha256=ND56BgUp_yLDTak2RsD9OqZK-X93eDGkL9Q97C2_Fz0,2191
|
|
4
|
+
pylocuszoom/coloc_plotter.py,sha256=9q76_4mYLNU6PjVw_rqx2THdTleI9U6mpzGBA6PnfrY,14036
|
|
5
|
+
pylocuszoom/colors.py,sha256=j1qBHHNTIE9k4bAkIrl7B7bTpTQdWq34-DdJ_y7hZPk,8751
|
|
6
|
+
pylocuszoom/config.py,sha256=Ow8YiW5Yl801K71jOnrPivHvAbA58uH1BvCAbV9hojM,16008
|
|
5
7
|
pylocuszoom/ensembl.py,sha256=w2msgBoIrY79iHI3hURSbevvdFHxHyWF9Z78hXtAaBc,14296
|
|
6
8
|
pylocuszoom/eqtl.py,sha256=9hGcFARABQRCMN3rco0pVlFJdmlh4SLBBKSgOvdIH_U,5924
|
|
7
9
|
pylocuszoom/exceptions.py,sha256=nd-rWMUodW62WVV4TfcYVPQcb66xV6v9FA-_4xHb5VY,926
|
|
8
10
|
pylocuszoom/finemapping.py,sha256=S3ulQj3fkaDM3n4I8EBymbWym_kTD5NEqfIEj93Mdjk,9630
|
|
9
11
|
pylocuszoom/forest.py,sha256=K-wBinxBOqIzsNMtZJ587e_oMhUXIXEqmEzVTUbmHSY,1161
|
|
10
12
|
pylocuszoom/gene_track.py,sha256=Sh0JCSdLNAAH0NQEiDVMvyXjm63PiCMq3gLvewcagvo,17277
|
|
11
|
-
pylocuszoom/labels.py,sha256=
|
|
12
|
-
pylocuszoom/ld.py,sha256=
|
|
13
|
+
pylocuszoom/labels.py,sha256=RPgEcCGPA2VQ1rXIW3WYy0AQ0EE0VBzbh_4EkhsWsNc,4343
|
|
14
|
+
pylocuszoom/ld.py,sha256=CphRau26XL9MoHiU3qRXlSz1Eg39TJt7MNIPZSZDr8M,13999
|
|
15
|
+
pylocuszoom/ld_heatmap_plotter.py,sha256=siHKL-tq7qQBTgyfH09uL0YJPmOGxG3udq8p1We5p0I,8373
|
|
13
16
|
pylocuszoom/loaders.py,sha256=KpWPBO0BCb2yrGTtgdiOqOuhx2YLmjK_ywmpr3onnx8,25156
|
|
14
17
|
pylocuszoom/logging.py,sha256=nZHEkbnjp8zoyWj_S-Hy9UQvUYLoMoxyiOWRozBT2dg,4987
|
|
15
18
|
pylocuszoom/manhattan.py,sha256=sNhPnsfsIqe1ls74D-kKMFyF_ZmaYB9Ul8qf4UMWnF0,8022
|
|
16
19
|
pylocuszoom/manhattan_plotter.py,sha256=1QQxaXEh5YG4x6ZIxpdhdfQPI2KuO_525qYKI7c32n4,27584
|
|
20
|
+
pylocuszoom/miami_plotter.py,sha256=J7GcgeIKyJvJTpQQHx9YNwH5NzpYSej2HPFSGu5YeLY,18099
|
|
17
21
|
pylocuszoom/phewas.py,sha256=6g2LmwA5kmxYlHgPxJvuXIMerEqfqgsrth110Y3CgVU,968
|
|
18
|
-
pylocuszoom/plotter.py,sha256=
|
|
22
|
+
pylocuszoom/plotter.py,sha256=I6fktn1mwE-ZM6bnKUqXrdN_eMQ6oHGaJBq9DwoT4xI,60488
|
|
19
23
|
pylocuszoom/py.typed,sha256=47DEQpj8HBSa-_TImW-5JCeuQeRkm5NMpJWZG3hSuFU,0
|
|
20
24
|
pylocuszoom/qq.py,sha256=GPIFHXYCLvhP4IUgjcU3QELLREH8r1AEYXMord8gtEo,3650
|
|
21
25
|
pylocuszoom/recombination.py,sha256=M-wDBdGbC5qGDHPFoGzBTPmTdiRD7bpRskyxBAKxTUY,15878
|
|
@@ -24,13 +28,13 @@ pylocuszoom/stats_plotter.py,sha256=67bgU-TXGnmVTxfTRWT3-PFemVVy6lTu4-ZlxUnwHS4,
|
|
|
24
28
|
pylocuszoom/utils.py,sha256=Z2P__Eau3ilF2ftuAZBm11EZ1NqCFQzfr4br9jCiJmg,6887
|
|
25
29
|
pylocuszoom/validation.py,sha256=3D9axjUvNXWW3Mk7dwRG38-di2P0zDpVVGF5WNSfZbk,7403
|
|
26
30
|
pylocuszoom/backends/__init__.py,sha256=xefVj3jVxmYwVLLY5AZtFqTPMehQxZ2qGd-Pk7_V_Bk,4267
|
|
27
|
-
pylocuszoom/backends/base.py,sha256=
|
|
28
|
-
pylocuszoom/backends/bokeh_backend.py,sha256=
|
|
31
|
+
pylocuszoom/backends/base.py,sha256=Wgzr6ncKnqZpjEYrc6aPmIKDWdZLm18mMO7q7XRU6SA,25640
|
|
32
|
+
pylocuszoom/backends/bokeh_backend.py,sha256=ft0W5ScxPoA6T6GB7k9PFfTMgOCyFom_plqVItdaUA0,34198
|
|
29
33
|
pylocuszoom/backends/hover.py,sha256=Hjm_jcxJL8dDxO_Ye7jeWAUcHKlbH6oO8ZfGJ2MzIFM,6564
|
|
30
|
-
pylocuszoom/backends/matplotlib_backend.py,sha256=
|
|
31
|
-
pylocuszoom/backends/plotly_backend.py,sha256=
|
|
34
|
+
pylocuszoom/backends/matplotlib_backend.py,sha256=SKTL_6QCc2D_UVITH543O-zPD_I8AwDLCZE0Re0KfE0,27269
|
|
35
|
+
pylocuszoom/backends/plotly_backend.py,sha256=cow67G8dIcrxSI3XOPFWJ9hSLqe1-MB4NFSeO8GDRG4,42228
|
|
32
36
|
pylocuszoom/reference_data/__init__.py,sha256=qqHqAUt1jebGlCN3CjqW3Z-_coHVNo5K3a3bb9o83hA,109
|
|
33
|
-
pylocuszoom-1.
|
|
34
|
-
pylocuszoom-1.
|
|
35
|
-
pylocuszoom-1.
|
|
36
|
-
pylocuszoom-1.
|
|
37
|
+
pylocuszoom-1.3.1.dist-info/METADATA,sha256=nHe6YBAHx718EndDz71xIukqf_WwJ7Q4yERBkgz9xmo,27020
|
|
38
|
+
pylocuszoom-1.3.1.dist-info/WHEEL,sha256=WLgqFyCfm_KASv4WHyYy0P3pM_m7J5L9k2skdKLirC8,87
|
|
39
|
+
pylocuszoom-1.3.1.dist-info/licenses/LICENSE.md,sha256=bqhD2fIhoqfLdX1lyGNoufIHWE7Q8mU-SyFadwKn4cc,34902
|
|
40
|
+
pylocuszoom-1.3.1.dist-info/RECORD,,
|