biopipen 0.28.1__py3-none-any.whl → 0.29.1__py3-none-any.whl
This diff represents the content of publicly available package versions that have been released to one of the supported registries. The information contained in this diff is provided for informational purposes only and reflects changes between package versions as they appear in their respective public registries.
Potentially problematic release.
This version of biopipen might be problematic. Click here for more details.
- biopipen/__init__.py +1 -1
- biopipen/core/config.toml +8 -0
- biopipen/ns/bam.py +0 -2
- biopipen/ns/bed.py +35 -0
- biopipen/ns/cellranger_pipeline.py +5 -5
- biopipen/ns/cnv.py +18 -2
- biopipen/ns/cnvkit_pipeline.py +16 -11
- biopipen/ns/gene.py +68 -23
- biopipen/ns/misc.py +2 -15
- biopipen/ns/plot.py +204 -0
- biopipen/ns/regulatory.py +214 -0
- biopipen/ns/scrna.py +31 -5
- biopipen/ns/snp.py +516 -8
- biopipen/ns/stats.py +167 -3
- biopipen/ns/vcf.py +196 -0
- biopipen/reports/snp/PlinkCallRate.svelte +24 -0
- biopipen/reports/snp/PlinkFreq.svelte +18 -0
- biopipen/reports/snp/PlinkHWE.svelte +18 -0
- biopipen/reports/snp/PlinkHet.svelte +18 -0
- biopipen/reports/snp/PlinkIBD.svelte +18 -0
- biopipen/scripts/bam/CNVpytor.py +144 -46
- biopipen/scripts/bed/BedtoolsIntersect.py +54 -0
- biopipen/scripts/bed/BedtoolsMerge.py +1 -1
- biopipen/scripts/cnv/AneuploidyScore.R +30 -7
- biopipen/scripts/cnv/AneuploidyScoreSummary.R +5 -2
- biopipen/scripts/cnv/TMADScore.R +21 -5
- biopipen/scripts/cnv/TMADScoreSummary.R +6 -2
- biopipen/scripts/cnvkit/CNVkitAccess.py +2 -1
- biopipen/scripts/cnvkit/CNVkitAutobin.py +3 -2
- biopipen/scripts/cnvkit/CNVkitBatch.py +1 -1
- biopipen/scripts/cnvkit/CNVkitCoverage.py +2 -1
- biopipen/scripts/cnvkit/CNVkitGuessBaits.py +1 -1
- biopipen/scripts/cnvkit/CNVkitHeatmap.py +1 -1
- biopipen/scripts/cnvkit/CNVkitReference.py +2 -1
- biopipen/scripts/delim/SampleInfo.R +10 -5
- biopipen/scripts/gene/GeneNameConversion.R +65 -0
- biopipen/scripts/gene/GenePromoters.R +61 -0
- biopipen/scripts/misc/Shell.sh +15 -0
- biopipen/scripts/plot/Manhattan.R +146 -0
- biopipen/scripts/plot/QQPlot.R +146 -0
- biopipen/scripts/regulatory/MotifAffinityTest.R +226 -0
- biopipen/scripts/regulatory/MotifAffinityTest_AtSNP.R +126 -0
- biopipen/scripts/regulatory/MotifAffinityTest_MotifBreakR.R +96 -0
- biopipen/scripts/regulatory/MotifScan.py +159 -0
- biopipen/scripts/regulatory/atSNP.R +33 -0
- biopipen/scripts/regulatory/motifBreakR.R +1594 -0
- biopipen/scripts/scrna/MarkersFinder.R +69 -67
- biopipen/scripts/scrna/SeuratClustering.R +71 -29
- biopipen/scripts/scrna/SeuratMap2Ref.R +20 -0
- biopipen/scripts/scrna/SeuratPreparing.R +252 -122
- biopipen/scripts/scrna/SeuratSubClustering.R +76 -27
- biopipen/scripts/snp/MatrixEQTL.R +85 -44
- biopipen/scripts/snp/Plink2GTMat.py +133 -0
- biopipen/scripts/snp/PlinkCallRate.R +190 -0
- biopipen/scripts/snp/PlinkFilter.py +100 -0
- biopipen/scripts/snp/PlinkFreq.R +298 -0
- biopipen/scripts/snp/PlinkFromVcf.py +78 -0
- biopipen/scripts/snp/PlinkHWE.R +80 -0
- biopipen/scripts/snp/PlinkHet.R +92 -0
- biopipen/scripts/snp/PlinkIBD.R +200 -0
- biopipen/scripts/snp/PlinkUpdateName.py +124 -0
- biopipen/scripts/stats/Mediation.R +94 -0
- biopipen/scripts/stats/MetaPvalue.R +2 -1
- biopipen/scripts/stats/MetaPvalue1.R +70 -0
- biopipen/scripts/tcr/TCRClusterStats.R +12 -7
- biopipen/scripts/vcf/BcftoolsAnnotate.py +91 -0
- biopipen/scripts/vcf/BcftoolsFilter.py +90 -0
- biopipen/scripts/vcf/BcftoolsSort.py +113 -0
- biopipen/scripts/vcf/BcftoolsView.py +73 -0
- biopipen/scripts/vcf/VcfFix_utils.py +1 -1
- biopipen/scripts/vcf/bcftools_utils.py +52 -0
- biopipen/utils/gene.R +83 -37
- biopipen/utils/gene.py +108 -60
- biopipen/utils/misc.R +56 -0
- biopipen/utils/misc.py +5 -2
- biopipen/utils/reference.py +54 -10
- {biopipen-0.28.1.dist-info → biopipen-0.29.1.dist-info}/METADATA +2 -2
- {biopipen-0.28.1.dist-info → biopipen-0.29.1.dist-info}/RECORD +80 -51
- {biopipen-0.28.1.dist-info → biopipen-0.29.1.dist-info}/entry_points.txt +1 -1
- biopipen/ns/bcftools.py +0 -111
- biopipen/scripts/bcftools/BcftoolsAnnotate.py +0 -42
- biopipen/scripts/bcftools/BcftoolsFilter.py +0 -79
- biopipen/scripts/bcftools/BcftoolsSort.py +0 -19
- biopipen/scripts/gene/GeneNameConversion.py +0 -66
- {biopipen-0.28.1.dist-info → biopipen-0.29.1.dist-info}/WHEEL +0 -0
biopipen/ns/stats.py
CHANGED
|
@@ -73,11 +73,103 @@ class ChowTest(Proc):
|
|
|
73
73
|
script = "file://../scripts/stats/ChowTest.R"
|
|
74
74
|
|
|
75
75
|
|
|
76
|
+
class Mediation(Proc):
|
|
77
|
+
"""Mediation analysis.
|
|
78
|
+
|
|
79
|
+
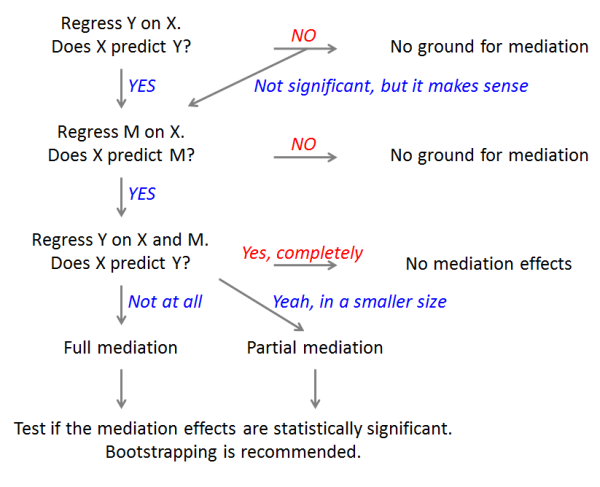
The flowchart of mediation analysis:
|
|
80
|
+
|
|
81
|
+
|
|
82
|
+
|
|
83
|
+
Reference:
|
|
84
|
+
- <https://library.virginia.edu/data/articles/introduction-to-mediation-analysis>
|
|
85
|
+
- <https://en.wikipedia.org/wiki/Mediation_(statistics)>
|
|
86
|
+
- <https://tilburgsciencehub.com/topics/analyze/regression/linear-regression/mediation-analysis/>
|
|
87
|
+
- <https://ademos.people.uic.edu/Chapter14.html>
|
|
88
|
+
|
|
89
|
+
Input:
|
|
90
|
+
infile: The input data file. The rows are samples and the columns are
|
|
91
|
+
features. It must be tab-delimited.
|
|
92
|
+
```
|
|
93
|
+
Sample F1 F2 F3 ... Fn
|
|
94
|
+
S1 1.2 3.4 5.6 7.8
|
|
95
|
+
S2 2.3 4.5 6.7 8.9
|
|
96
|
+
...
|
|
97
|
+
Sm 5.6 7.8 9.0 1.2
|
|
98
|
+
```
|
|
99
|
+
fmlfile: The formula file.
|
|
100
|
+
```
|
|
101
|
+
Case M Y X Cov Model_M Model_Y
|
|
102
|
+
Case1 F1 F2 F3 F4,F5 glm lm
|
|
103
|
+
...
|
|
104
|
+
```
|
|
105
|
+
Where Y is the outcome variable, X is the predictor variable, M is the
|
|
106
|
+
mediator variable, and Case is the case name. Model_M and Model_Y are the
|
|
107
|
+
models for M and Y, respectively.
|
|
108
|
+
`envs.cases` will be ignored if this is provided.
|
|
109
|
+
|
|
110
|
+
Output:
|
|
111
|
+
outfile: The output file.
|
|
112
|
+
Columns to help understand the results:
|
|
113
|
+
Total Effect: a total effect of X on Y (without M) (`Y ~ X`).
|
|
114
|
+
ADE: A Direct Effect of X on Y after taking into account a mediation effect of M (`Y ~ X + M`).
|
|
115
|
+
ACME: The Mediation Effect, the total effect minus the direct effect,
|
|
116
|
+
which equals to a product of a coefficient of X in the second step and a coefficient of M in the last step.
|
|
117
|
+
The goal of mediation analysis is to obtain this indirect effect and see if it's statistically significant.
|
|
118
|
+
|
|
119
|
+
Envs:
|
|
120
|
+
ncores (type=int): Number of cores to use for parallelization for cases.
|
|
121
|
+
sims (type=int): Number of Monte Carlo draws for nonparametric bootstrap or quasi-Bayesian approximation.
|
|
122
|
+
Will be passed to `mediation::mediate` function.
|
|
123
|
+
args (ns): Other arguments passed to `mediation::mediate` function.
|
|
124
|
+
- <more>: More arguments passed to `mediation::mediate` function.
|
|
125
|
+
See: <https://rdrr.io/cran/mediation/man/mediate.html>
|
|
126
|
+
padj (choice): The method for (ACME) p-value adjustment.
|
|
127
|
+
- none: No p-value adjustment (no Padj column in outfile).
|
|
128
|
+
- holm: Holm-Bonferroni method.
|
|
129
|
+
- hochberg: Hochberg method.
|
|
130
|
+
- hommel: Hommel method.
|
|
131
|
+
- bonferroni: Bonferroni method.
|
|
132
|
+
- BH: Benjamini-Hochberg method.
|
|
133
|
+
- BY: Benjamini-Yekutieli method.
|
|
134
|
+
- fdr: FDR correction method.
|
|
135
|
+
cases (type=json): The cases for mediation analysis.
|
|
136
|
+
Ignored if `in.fmlfile` is provided.
|
|
137
|
+
A json/dict with case names as keys and values as a dict of M, Y, X, Cov, Model_M, Model_Y.
|
|
138
|
+
For example:
|
|
139
|
+
```json
|
|
140
|
+
{
|
|
141
|
+
"Case1": {
|
|
142
|
+
"M": "F1",
|
|
143
|
+
"Y": "F2",
|
|
144
|
+
"X": "F3",
|
|
145
|
+
"Cov": "F4,F5",
|
|
146
|
+
"Model_M": "glm",
|
|
147
|
+
"Model_Y": "lm"
|
|
148
|
+
},
|
|
149
|
+
...
|
|
150
|
+
}
|
|
151
|
+
```
|
|
152
|
+
transpose_input (flag): Whether to transpose the input file.
|
|
153
|
+
""" # noqa: E501
|
|
154
|
+
input = "infile:file, fmlfile:file"
|
|
155
|
+
output = "outfile:file:{{in.infile | stem}}.mediation.txt"
|
|
156
|
+
lang = config.lang.rscript
|
|
157
|
+
envs = {
|
|
158
|
+
"ncores": config.misc.ncores,
|
|
159
|
+
"sims": 1000,
|
|
160
|
+
"args": {},
|
|
161
|
+
"padj": "none",
|
|
162
|
+
"cases": {},
|
|
163
|
+
"transpose_input": False,
|
|
164
|
+
}
|
|
165
|
+
script = "file://../scripts/stats/Mediation.R"
|
|
166
|
+
|
|
167
|
+
|
|
76
168
|
class LiquidAssoc(Proc):
|
|
77
169
|
"""Liquid association tests.
|
|
78
170
|
|
|
79
171
|
See Also https://github.com/gundt/fastLiquidAssociation
|
|
80
|
-
|
|
172
|
+
Requires https://github.com/pwwang/fastLiquidAssociation
|
|
81
173
|
|
|
82
174
|
Input:
|
|
83
175
|
infile: The input data file. The rows are samples and the columns are
|
|
@@ -275,8 +367,10 @@ class MetaPvalue(Proc):
|
|
|
275
367
|
|
|
276
368
|
Envs:
|
|
277
369
|
id_cols: The column names used in all `in.infiles` as ID columns. Multiple
|
|
278
|
-
columns can be specified by comma-seperated values. For example, `ID1,ID2
|
|
279
|
-
|
|
370
|
+
columns can be specified by comma-seperated values. For example, `ID1,ID2`,
|
|
371
|
+
where `ID1` is the ID column in the first file and `ID2` is the ID column
|
|
372
|
+
in the second file.
|
|
373
|
+
If `id_exprs` is specified, this should be a single column name for the new
|
|
280
374
|
ID column in each `in.infiles` and the final `out.outfile`.
|
|
281
375
|
id_exprs: The R expressions for each `in.infiles` to get ID column(s).
|
|
282
376
|
pval_cols: The column names used in all `in.infiles` as p-value columns.
|
|
@@ -294,6 +388,8 @@ class MetaPvalue(Proc):
|
|
|
294
388
|
- votep: Vote counting method.
|
|
295
389
|
- wilkinsonp: Wilkinson's method.
|
|
296
390
|
- invchisq: Inverse chi-square method.
|
|
391
|
+
keep_single (flag): Whether to keep the original p-value when there is only one
|
|
392
|
+
p-value.
|
|
297
393
|
na: The method to handle NA values. -1 to skip the record. Otherwise NA
|
|
298
394
|
will be replaced by the given value.
|
|
299
395
|
padj (choice): The method for p-value adjustment.
|
|
@@ -315,6 +411,74 @@ class MetaPvalue(Proc):
|
|
|
315
411
|
"pval_cols": None,
|
|
316
412
|
"method": "fisher",
|
|
317
413
|
"na": -1,
|
|
414
|
+
"keep_single": True,
|
|
318
415
|
"padj": "none",
|
|
319
416
|
}
|
|
320
417
|
script = "file://../scripts/stats/MetaPvalue.R"
|
|
418
|
+
|
|
419
|
+
|
|
420
|
+
class MetaPvalue1(Proc):
|
|
421
|
+
"""Calulation of meta p-values.
|
|
422
|
+
|
|
423
|
+
Unlike `MetaPvalue`, this process only accepts one input file.
|
|
424
|
+
|
|
425
|
+
The p-values will be grouped by the ID columns and combined by the selected method.
|
|
426
|
+
|
|
427
|
+
Input:
|
|
428
|
+
infile: The input file.
|
|
429
|
+
The file is a tab-delimited file with multiple
|
|
430
|
+
columns. There should be ID column(s) to group the rows where
|
|
431
|
+
p-value column(s) to be combined.
|
|
432
|
+
|
|
433
|
+
Output:
|
|
434
|
+
outfile: The output file. It is a tab-delimited file with the first column as
|
|
435
|
+
the ID and the second column as the combined p-value.
|
|
436
|
+
```
|
|
437
|
+
ID ID1 ... Pval Padj
|
|
438
|
+
a x ... 0.123 0.123
|
|
439
|
+
b y ... 0.123 0.123
|
|
440
|
+
...
|
|
441
|
+
```
|
|
442
|
+
|
|
443
|
+
Envs:
|
|
444
|
+
id_cols: The column names used in `in.infile` as ID columns. Multiple
|
|
445
|
+
columns can be specified by comma-seperated values. For example, `ID1,ID2`.
|
|
446
|
+
pval_col: The column name used in `in.infile` as p-value column.
|
|
447
|
+
method (choice): The method used to calculate the meta-pvalue.
|
|
448
|
+
- fisher: Fisher's method.
|
|
449
|
+
- sumlog: Sum of logarithms (same as Fisher's method)
|
|
450
|
+
- logitp: Logit method.
|
|
451
|
+
- sumz: Sum of z method (Stouffer's method).
|
|
452
|
+
- meanz: Mean of z method.
|
|
453
|
+
- meanp: Mean of p method.
|
|
454
|
+
- invt: Inverse t method.
|
|
455
|
+
- sump: Sum of p method (Edgington's method).
|
|
456
|
+
- votep: Vote counting method.
|
|
457
|
+
- wilkinsonp: Wilkinson's method.
|
|
458
|
+
- invchisq: Inverse chi-square method.
|
|
459
|
+
na: The method to handle NA values. -1 to skip the record. Otherwise NA
|
|
460
|
+
will be replaced by the given value.
|
|
461
|
+
keep_single (flag): Whether to keep the original p-value when there is only one
|
|
462
|
+
p-value.
|
|
463
|
+
padj (choice): The method for p-value adjustment.
|
|
464
|
+
- none: No p-value adjustment (no Padj column in outfile).
|
|
465
|
+
- holm: Holm-Bonferroni method.
|
|
466
|
+
- hochberg: Hochberg method.
|
|
467
|
+
- hommel: Hommel method.
|
|
468
|
+
- bonferroni: Bonferroni method.
|
|
469
|
+
- BH: Benjamini-Hochberg method.
|
|
470
|
+
- BY: Benjamini-Yekutieli method.
|
|
471
|
+
- fdr: FDR correction method.
|
|
472
|
+
"""
|
|
473
|
+
input = "infile:file"
|
|
474
|
+
output = "outfile:file:{{in.infile | stem}}.metapval.txt"
|
|
475
|
+
lang = config.lang.rscript
|
|
476
|
+
envs = {
|
|
477
|
+
"id_cols": None,
|
|
478
|
+
"pval_col": None,
|
|
479
|
+
"method": "fisher",
|
|
480
|
+
"na": -1,
|
|
481
|
+
"keep_single": True,
|
|
482
|
+
"padj": "none",
|
|
483
|
+
}
|
|
484
|
+
script = "file://../scripts/stats/MetaPvalue1.R"
|
biopipen/ns/vcf.py
CHANGED
|
@@ -439,3 +439,199 @@ class TruvariConsistency(Proc):
|
|
|
439
439
|
envs = {"truvari": config.exe.truvari, "heatmap": {}}
|
|
440
440
|
script = "file://../scripts/vcf/TruvariConsistency.R"
|
|
441
441
|
plugin_opts = {"report": "file://../reports/vcf/TruvariConsistency.svelte"}
|
|
442
|
+
|
|
443
|
+
|
|
444
|
+
class BcftoolsAnnotate(Proc):
|
|
445
|
+
"""Add or remove annotations from VCF files
|
|
446
|
+
|
|
447
|
+
See also: <https://samtools.github.io/bcftools/bcftools.html#annotate>
|
|
448
|
+
|
|
449
|
+
Input:
|
|
450
|
+
infile: The input VCF file
|
|
451
|
+
annfile: The annotation file.
|
|
452
|
+
Currently only VCF files are supported.
|
|
453
|
+
|
|
454
|
+
Output:
|
|
455
|
+
outfile: The VCF file with annotations added or removed.
|
|
456
|
+
|
|
457
|
+
Envs:
|
|
458
|
+
bcftools: Path to bcftools
|
|
459
|
+
tabix: Path to tabix, used to index infile and annfile
|
|
460
|
+
annfile: The annotation file. If `in.annfile` is provided,
|
|
461
|
+
this is ignored
|
|
462
|
+
ncores (type=int): Number of cores (`--threads`) to use
|
|
463
|
+
columns (auto): Comma-separated or list of columns or tags to carry over from
|
|
464
|
+
the annotation file. Overrides `-c, --columns`
|
|
465
|
+
remove (auto): Remove the specified columns from the input file
|
|
466
|
+
header (type=list): Headers to be added
|
|
467
|
+
gz (flag): Whether to gzip the output file
|
|
468
|
+
index (flag): Whether to index the output file (tbi) (`envs.gz` forced to True)
|
|
469
|
+
<more>: Other arguments for `bcftools annotate`
|
|
470
|
+
See also <https://samtools.github.io/bcftools/bcftools.html#annotate>
|
|
471
|
+
Note that the underscore `_` will be replaced with dash `-` in the
|
|
472
|
+
argument name.
|
|
473
|
+
"""
|
|
474
|
+
input = "infile:file, annfile:file"
|
|
475
|
+
output = (
|
|
476
|
+
"outfile:file:{{in.infile | stem: 'gz'}}.vcf"
|
|
477
|
+
"{{'.gz' if envs.index or envs.gz else ''}}"
|
|
478
|
+
)
|
|
479
|
+
lang = config.lang.python
|
|
480
|
+
envs = {
|
|
481
|
+
"bcftools": config.exe.bcftools,
|
|
482
|
+
"tabix": config.exe.tabix,
|
|
483
|
+
"annfile": None,
|
|
484
|
+
"columns": [],
|
|
485
|
+
"remove": [],
|
|
486
|
+
"header": [],
|
|
487
|
+
"gz": True,
|
|
488
|
+
"index": True,
|
|
489
|
+
"ncores": config.misc.ncores,
|
|
490
|
+
}
|
|
491
|
+
script = "file://../scripts/vcf/BcftoolsAnnotate.py"
|
|
492
|
+
|
|
493
|
+
|
|
494
|
+
class BcftoolsFilter(Proc):
|
|
495
|
+
"""Apply fixed threshold filters to VCF files
|
|
496
|
+
|
|
497
|
+
Input:
|
|
498
|
+
infile: The input VCF file
|
|
499
|
+
|
|
500
|
+
Output:
|
|
501
|
+
outfile: The filtered VCF file. If the `in.infile` is gzipped, this is
|
|
502
|
+
gzipped as well.
|
|
503
|
+
|
|
504
|
+
Envs:
|
|
505
|
+
bcftools: Path to bcftools
|
|
506
|
+
tabix: Path to tabix, used to index infile/outfile
|
|
507
|
+
ncores (type=int): Number of cores (`--threads`) to use
|
|
508
|
+
keep: Whether we should keep the filtered variants or not.
|
|
509
|
+
If True, the filtered variants will be kept in the output file, but
|
|
510
|
+
with a new FILTER.
|
|
511
|
+
includes: and
|
|
512
|
+
excludes: include/exclude only sites for which EXPRESSION is true.
|
|
513
|
+
See: <https://samtools.github.io/bcftools/bcftools.html#expressions>
|
|
514
|
+
If provided, `envs.include/exclude` will be ignored.
|
|
515
|
+
If `str`/`list` used, The filter names will be `Filter_<type>_<index>`.
|
|
516
|
+
A dict is used where keys are filter names and values are expressions
|
|
517
|
+
gz (flag): Whether to gzip the output file
|
|
518
|
+
index (flag): Whether to index the output file (tbi) (`envs.gz` forced to True)
|
|
519
|
+
<more>: Other arguments for `bcftools filter`
|
|
520
|
+
See also <https://samtools.github.io/bcftools/bcftools.html#filter>
|
|
521
|
+
"""
|
|
522
|
+
input = "infile:file"
|
|
523
|
+
output = (
|
|
524
|
+
"outfile:file:{{in.infile | stem: 'gz'}}.vcf"
|
|
525
|
+
"{{'.gz' if envs.index or envs.gz else ''}}"
|
|
526
|
+
)
|
|
527
|
+
lang = config.lang.python
|
|
528
|
+
envs = {
|
|
529
|
+
"bcftools": config.exe.bcftools,
|
|
530
|
+
"tabix": config.exe.tabix,
|
|
531
|
+
"ncores": config.misc.ncores,
|
|
532
|
+
"keep": True,
|
|
533
|
+
"includes": None,
|
|
534
|
+
"excludes": None,
|
|
535
|
+
"gz": True,
|
|
536
|
+
"index": True,
|
|
537
|
+
}
|
|
538
|
+
script = "file://../scripts/vcf/BcftoolsFilter.py"
|
|
539
|
+
|
|
540
|
+
|
|
541
|
+
class BcftoolsSort(Proc):
|
|
542
|
+
"""Sort VCF files using `bcftools sort`.
|
|
543
|
+
|
|
544
|
+
`bcftools sort` is used to sort VCF files by chromosome and position based on the
|
|
545
|
+
order of contigs in the header.
|
|
546
|
+
|
|
547
|
+
Here we provide a chrsize file to first sort the contigs in the header and then
|
|
548
|
+
sort the VCF file using `bcftools sort`.
|
|
549
|
+
|
|
550
|
+
Input:
|
|
551
|
+
infile: The input VCF file
|
|
552
|
+
|
|
553
|
+
Output:
|
|
554
|
+
outfile: The sorted VCF file.
|
|
555
|
+
|
|
556
|
+
Envs:
|
|
557
|
+
bcftools: Path to bcftools
|
|
558
|
+
tabix: Path to tabix, used to index infile/outfile
|
|
559
|
+
ncores (type=int): Number of cores (`--threads`) to use
|
|
560
|
+
gz (flag): Whether to gzip the output file
|
|
561
|
+
index (flag): Whether to index the output file (tbi) (`envs.gz` forced to True)
|
|
562
|
+
chrsize: The chromosome size file, from which the chromosome order is used
|
|
563
|
+
to sort the contig in the header first.
|
|
564
|
+
If not provided, `bcftools sort` will be used directly.
|
|
565
|
+
notfound (choice): What if the contig in the VCF file is not found in the
|
|
566
|
+
`chrsize` file.
|
|
567
|
+
- error: Report error
|
|
568
|
+
- remove: Remove the contig from the header.
|
|
569
|
+
Note that if there are records with the removed contig, an error will
|
|
570
|
+
be raised by `bcftools sort`
|
|
571
|
+
- start: Move the contig to the start of the contigs from `chrsize`
|
|
572
|
+
- end: Move the contig to the end of the contigs from `chrsize`
|
|
573
|
+
<more>: Other arguments for `bcftools sort`. For example `max_mem`.
|
|
574
|
+
See also <https://samtools.github.io/bcftools/bcftools.html#sort>
|
|
575
|
+
"""
|
|
576
|
+
input = "infile:file"
|
|
577
|
+
output = (
|
|
578
|
+
"outfile:file:{{in.infile | stem: 'gz'}}.vcf"
|
|
579
|
+
"{{'.gz' if envs.index or envs.gz else ''}}"
|
|
580
|
+
)
|
|
581
|
+
lang = config.lang.python
|
|
582
|
+
envs = {
|
|
583
|
+
"bcftools": config.exe.bcftools,
|
|
584
|
+
"tabix": config.exe.tabix,
|
|
585
|
+
"ncores": config.misc.ncores,
|
|
586
|
+
"chrsize": config.ref.chrsize,
|
|
587
|
+
"notfound": "remove",
|
|
588
|
+
"gz": True,
|
|
589
|
+
"index": True,
|
|
590
|
+
}
|
|
591
|
+
script = "file://../scripts/vcf/BcftoolsSort.py"
|
|
592
|
+
|
|
593
|
+
|
|
594
|
+
class BcftoolsView(Proc):
|
|
595
|
+
"""View, subset and filter VCF files by position and filtering expression.
|
|
596
|
+
|
|
597
|
+
Also convert between VCF and BCF.
|
|
598
|
+
|
|
599
|
+
Input:
|
|
600
|
+
infile: The input VCF file
|
|
601
|
+
regions_file: The region file used to subset the input VCF file.
|
|
602
|
+
samples_file: The samples file used to subset the input VCF file.
|
|
603
|
+
|
|
604
|
+
Output:
|
|
605
|
+
outfile: The output VCF file.
|
|
606
|
+
|
|
607
|
+
Envs:
|
|
608
|
+
bcftools: Path to bcftools
|
|
609
|
+
tabix: Path to tabix, used to index infile/outfile
|
|
610
|
+
ncores (type=int): Number of cores (`--threads`) to use
|
|
611
|
+
regions_file: The region file used to subset the input VCF file.
|
|
612
|
+
If `in.regions_file` is provided, this is ignored.
|
|
613
|
+
samples_file: The samples file used to subset the input VCF file.
|
|
614
|
+
If `in.samples_file` is provided, this is ignored.
|
|
615
|
+
gz (flag): Whether to gzip the output file
|
|
616
|
+
index (flag): Whether to index the output file (tbi) (`envs.gz` forced to True)
|
|
617
|
+
<more>: Other arguments for `bcftools view`.
|
|
618
|
+
See also https://samtools.github.io/bcftools/bcftools.html#view
|
|
619
|
+
Note that the underscore `_` will be replaced with dash `-` in the
|
|
620
|
+
argument name.
|
|
621
|
+
"""
|
|
622
|
+
input = "infile:file, regions_file:file, samples_file:file"
|
|
623
|
+
output = (
|
|
624
|
+
"outfile:file:{{in.infile | stem: 'gz'}}.vcf"
|
|
625
|
+
"{{'.gz' if envs.index or envs.gz else ''}}"
|
|
626
|
+
)
|
|
627
|
+
lang = config.lang.python
|
|
628
|
+
envs = {
|
|
629
|
+
"bcftools": config.exe.bcftools,
|
|
630
|
+
"tabix": config.exe.tabix,
|
|
631
|
+
"ncores": config.misc.ncores,
|
|
632
|
+
"regions_file": None,
|
|
633
|
+
"samples_file": None,
|
|
634
|
+
"gz": True,
|
|
635
|
+
"index": True,
|
|
636
|
+
}
|
|
637
|
+
script = "file://../scripts/vcf/BcftoolsView.py"
|
|
@@ -0,0 +1,24 @@
|
|
|
1
|
+
{% from "utils/misc.liq" import report_jobs -%}
|
|
2
|
+
<script>
|
|
3
|
+
import { Image, Descr } from "$libs";
|
|
4
|
+
</script>
|
|
5
|
+
|
|
6
|
+
{%- macro report_job(job, h=1) -%}
|
|
7
|
+
<h{{h+1}}>Sample Call Rate</h{{h+1}}>
|
|
8
|
+
{%- for pngfile in job.out.outdir | joinpaths: '*.samplecr.png' | glob -%}
|
|
9
|
+
<Descr>Cutoff: {{envs.samplecr}}</Descr>
|
|
10
|
+
<Image src="{{pngfile}}" />
|
|
11
|
+
{%- endfor -%}
|
|
12
|
+
|
|
13
|
+
<h{{h+1}}>Variant Call Rate</h{{h+1}}>
|
|
14
|
+
{%- for pngfile in job.out.outdir | joinpaths: '*.varcr.png' | glob -%}
|
|
15
|
+
<Descr>Cutoff: {{envs.varcr}}</Descr>
|
|
16
|
+
<Image src="{{pngfile}}" />
|
|
17
|
+
{%- endfor -%}
|
|
18
|
+
{%- endmacro -%}
|
|
19
|
+
|
|
20
|
+
{%- macro head_job(job) -%}
|
|
21
|
+
<h1>Sample: {{job.in.cnrfile | stem0 }}</h1>
|
|
22
|
+
{%- endmacro -%}
|
|
23
|
+
|
|
24
|
+
{{ report_jobs(jobs, head_job, report_job) }}
|
|
@@ -0,0 +1,18 @@
|
|
|
1
|
+
{% from "utils/misc.liq" import report_jobs -%}
|
|
2
|
+
<script>
|
|
3
|
+
import { Image, Descr } from "$libs";
|
|
4
|
+
</script>
|
|
5
|
+
|
|
6
|
+
{%- macro report_job(job, h=1) -%}
|
|
7
|
+
{%- for pngfile in job.out.outdir | joinpaths: '*.png' | glob -%}
|
|
8
|
+
{% set metric_col = pngfile | stem | ext0 %}
|
|
9
|
+
<h{{h+1}}>{{metric_col}} distribution</h{{h+1}}>
|
|
10
|
+
<Image src="{{pngfile}}" />
|
|
11
|
+
{%- endfor -%}
|
|
12
|
+
{%- endmacro -%}
|
|
13
|
+
|
|
14
|
+
{%- macro head_job(job) -%}
|
|
15
|
+
<h1>Sample: {{job.in.cnrfile | stem0 }}</h1>
|
|
16
|
+
{%- endmacro -%}
|
|
17
|
+
|
|
18
|
+
{{ report_jobs(jobs, head_job, report_job) }}
|
|
@@ -0,0 +1,18 @@
|
|
|
1
|
+
{% from "utils/misc.liq" import report_jobs -%}
|
|
2
|
+
<script>
|
|
3
|
+
import { Image, Descr } from "$libs";
|
|
4
|
+
</script>
|
|
5
|
+
|
|
6
|
+
{%- macro report_job(job, h=1) -%}
|
|
7
|
+
{%- for pngfile in job.out.outdir | joinpaths: '*.png' | glob -%}
|
|
8
|
+
<h{{h+1}}>Distribution</h{{h+1}}>
|
|
9
|
+
<Descr>Cutoff: {{envs.cutoff}}</Descr>
|
|
10
|
+
<Image src="{{pngfile}}" />
|
|
11
|
+
{%- endfor -%}
|
|
12
|
+
{%- endmacro -%}
|
|
13
|
+
|
|
14
|
+
{%- macro head_job(job) -%}
|
|
15
|
+
<h1>Sample: {{job.in.cnrfile | stem0 }}</h1>
|
|
16
|
+
{%- endmacro -%}
|
|
17
|
+
|
|
18
|
+
{{ report_jobs(jobs, head_job, report_job) }}
|
|
@@ -0,0 +1,18 @@
|
|
|
1
|
+
{% from "utils/misc.liq" import report_jobs -%}
|
|
2
|
+
<script>
|
|
3
|
+
import { Image, Descr } from "$libs";
|
|
4
|
+
</script>
|
|
5
|
+
|
|
6
|
+
{%- macro report_job(job, h=1) -%}
|
|
7
|
+
{%- for pngfile in job.out.outdir | joinpaths: '*.png' | glob -%}
|
|
8
|
+
<h{{h+1}}>Distribution</h{{h+1}}>
|
|
9
|
+
<Descr>Cutoff: [mean - {{envs.cutoff}} x sd, mean + {{envs.cutoff}} x sd]</Descr>
|
|
10
|
+
<Image src="{{pngfile}}" />
|
|
11
|
+
{%- endfor -%}
|
|
12
|
+
{%- endmacro -%}
|
|
13
|
+
|
|
14
|
+
{%- macro head_job(job) -%}
|
|
15
|
+
<h1>Sample: {{job.in.cnrfile | stem0 }}</h1>
|
|
16
|
+
{%- endmacro -%}
|
|
17
|
+
|
|
18
|
+
{{ report_jobs(jobs, head_job, report_job) }}
|
|
@@ -0,0 +1,18 @@
|
|
|
1
|
+
{% from "utils/misc.liq" import report_jobs -%}
|
|
2
|
+
<script>
|
|
3
|
+
import { Image, Descr } from "$libs";
|
|
4
|
+
</script>
|
|
5
|
+
|
|
6
|
+
{%- macro report_job(job, h=1) -%}
|
|
7
|
+
{%- for pngfile in job.out.outdir | joinpaths: '*.png' | glob -%}
|
|
8
|
+
<h{{h+1}}>Heatmap</h{{h+1}}>
|
|
9
|
+
<Descr>PI_HAT threshold = {{envs.pihat}}</Descr>
|
|
10
|
+
<Image src="{{pngfile}}" />
|
|
11
|
+
{%- endfor -%}
|
|
12
|
+
{%- endmacro -%}
|
|
13
|
+
|
|
14
|
+
{%- macro head_job(job) -%}
|
|
15
|
+
<h1>Sample: {{job.in.cnrfile | stem0 }}</h1>
|
|
16
|
+
{%- endmacro -%}
|
|
17
|
+
|
|
18
|
+
{{ report_jobs(jobs, head_job, report_job) }}
|