advisor-scattering 0.5.0__py3-none-any.whl
This diff represents the content of publicly available package versions that have been released to one of the supported registries. The information contained in this diff is provided for informational purposes only and reflects changes between package versions as they appear in their respective public registries.
- advisor/__init__.py +3 -0
- advisor/__main__.py +7 -0
- advisor/app.py +40 -0
- advisor/controllers/__init__.py +6 -0
- advisor/controllers/app_controller.py +69 -0
- advisor/controllers/feature_controller.py +25 -0
- advisor/domain/__init__.py +23 -0
- advisor/domain/core/__init__.py +8 -0
- advisor/domain/core/lab.py +121 -0
- advisor/domain/core/lattice.py +79 -0
- advisor/domain/core/sample.py +101 -0
- advisor/domain/geometry.py +212 -0
- advisor/domain/unit_converter.py +82 -0
- advisor/features/__init__.py +6 -0
- advisor/features/scattering_geometry/controllers/__init__.py +5 -0
- advisor/features/scattering_geometry/controllers/scattering_geometry_controller.py +26 -0
- advisor/features/scattering_geometry/domain/__init__.py +5 -0
- advisor/features/scattering_geometry/domain/brillouin_calculator.py +410 -0
- advisor/features/scattering_geometry/domain/core.py +516 -0
- advisor/features/scattering_geometry/ui/__init__.py +5 -0
- advisor/features/scattering_geometry/ui/components/__init__.py +17 -0
- advisor/features/scattering_geometry/ui/components/angles_to_hkl_components.py +150 -0
- advisor/features/scattering_geometry/ui/components/hk_angles_components.py +430 -0
- advisor/features/scattering_geometry/ui/components/hkl_scan_components.py +526 -0
- advisor/features/scattering_geometry/ui/components/hkl_to_angles_components.py +315 -0
- advisor/features/scattering_geometry/ui/scattering_geometry_tab.py +725 -0
- advisor/features/structure_factor/controllers/__init__.py +6 -0
- advisor/features/structure_factor/controllers/structure_factor_controller.py +25 -0
- advisor/features/structure_factor/domain/__init__.py +6 -0
- advisor/features/structure_factor/domain/structure_factor_calculator.py +107 -0
- advisor/features/structure_factor/ui/__init__.py +6 -0
- advisor/features/structure_factor/ui/components/__init__.py +12 -0
- advisor/features/structure_factor/ui/components/customized_plane_components.py +358 -0
- advisor/features/structure_factor/ui/components/hkl_plane_components.py +391 -0
- advisor/features/structure_factor/ui/structure_factor_tab.py +273 -0
- advisor/resources/__init__.py +0 -0
- advisor/resources/config/app_config.json +14 -0
- advisor/resources/config/tips.json +4 -0
- advisor/resources/data/nacl.cif +111 -0
- advisor/resources/icons/bz_caculator.jpg +0 -0
- advisor/resources/icons/bz_calculator.png +0 -0
- advisor/resources/icons/minus.svg +3 -0
- advisor/resources/icons/placeholder.png +0 -0
- advisor/resources/icons/plus.svg +3 -0
- advisor/resources/icons/reset.png +0 -0
- advisor/resources/icons/sf_calculator.jpg +0 -0
- advisor/resources/icons/sf_calculator.png +0 -0
- advisor/resources/icons.qrc +6 -0
- advisor/resources/qss/styles.qss +348 -0
- advisor/resources/resources_rc.py +83 -0
- advisor/ui/__init__.py +7 -0
- advisor/ui/init_window.py +566 -0
- advisor/ui/main_window.py +174 -0
- advisor/ui/tab_interface.py +44 -0
- advisor/ui/tips.py +30 -0
- advisor/ui/utils/__init__.py +6 -0
- advisor/ui/utils/readcif.py +129 -0
- advisor/ui/visualizers/HKLScan2DVisualizer.py +224 -0
- advisor/ui/visualizers/__init__.py +8 -0
- advisor/ui/visualizers/coordinate_visualizer.py +203 -0
- advisor/ui/visualizers/scattering_visualizer.py +301 -0
- advisor/ui/visualizers/structure_factor_visualizer.py +426 -0
- advisor/ui/visualizers/structure_factor_visualizer_2d.py +235 -0
- advisor/ui/visualizers/unitcell_visualizer.py +518 -0
- advisor_scattering-0.5.0.dist-info/METADATA +122 -0
- advisor_scattering-0.5.0.dist-info/RECORD +69 -0
- advisor_scattering-0.5.0.dist-info/WHEEL +5 -0
- advisor_scattering-0.5.0.dist-info/entry_points.txt +3 -0
- advisor_scattering-0.5.0.dist-info/top_level.txt +1 -0
|
@@ -0,0 +1,518 @@
|
|
|
1
|
+
"""visualize the unit cell as done in VESTA."""
|
|
2
|
+
|
|
3
|
+
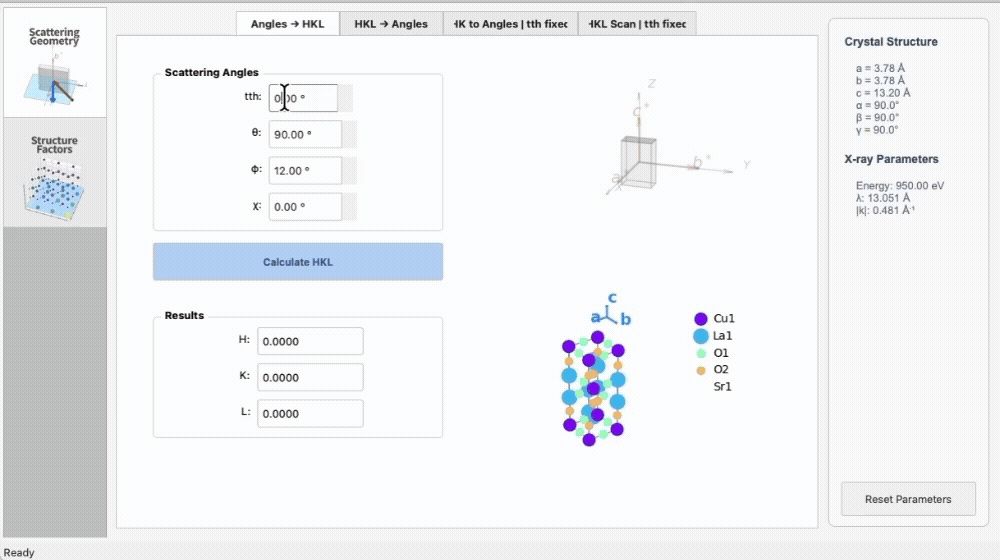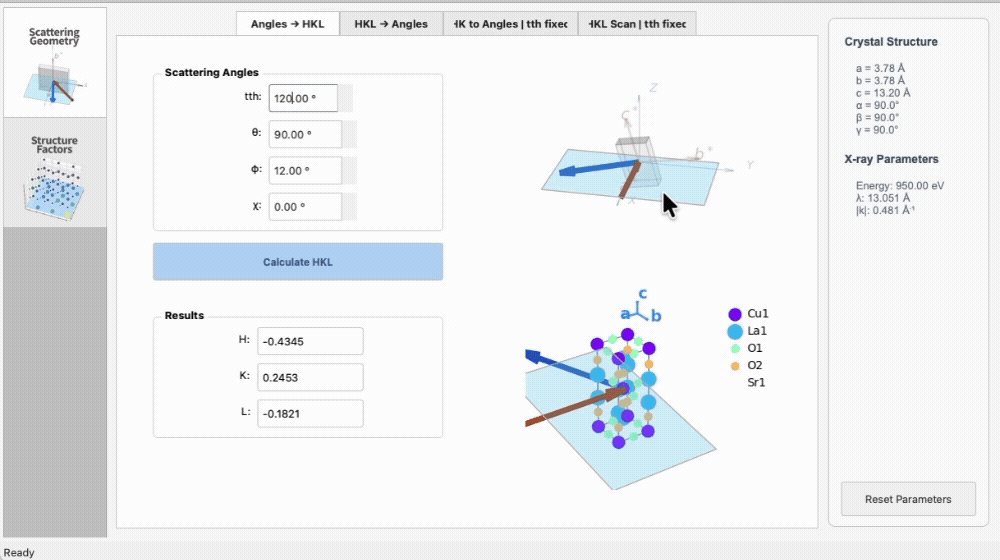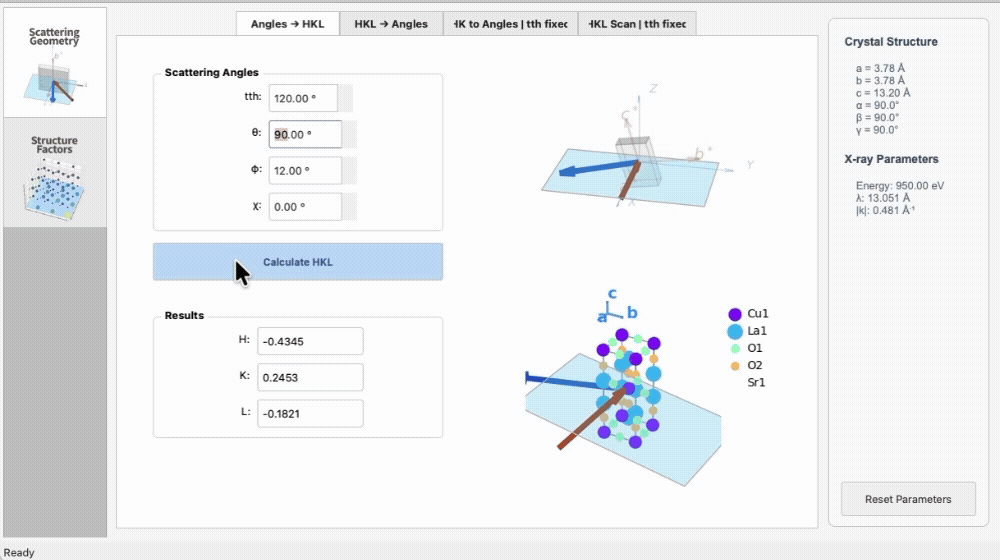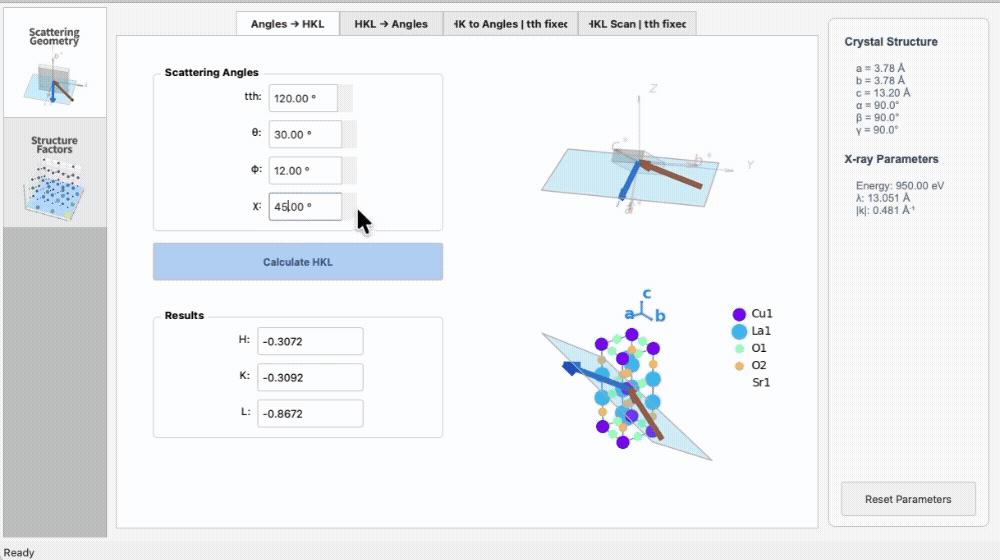
import os
|
|
4
|
+
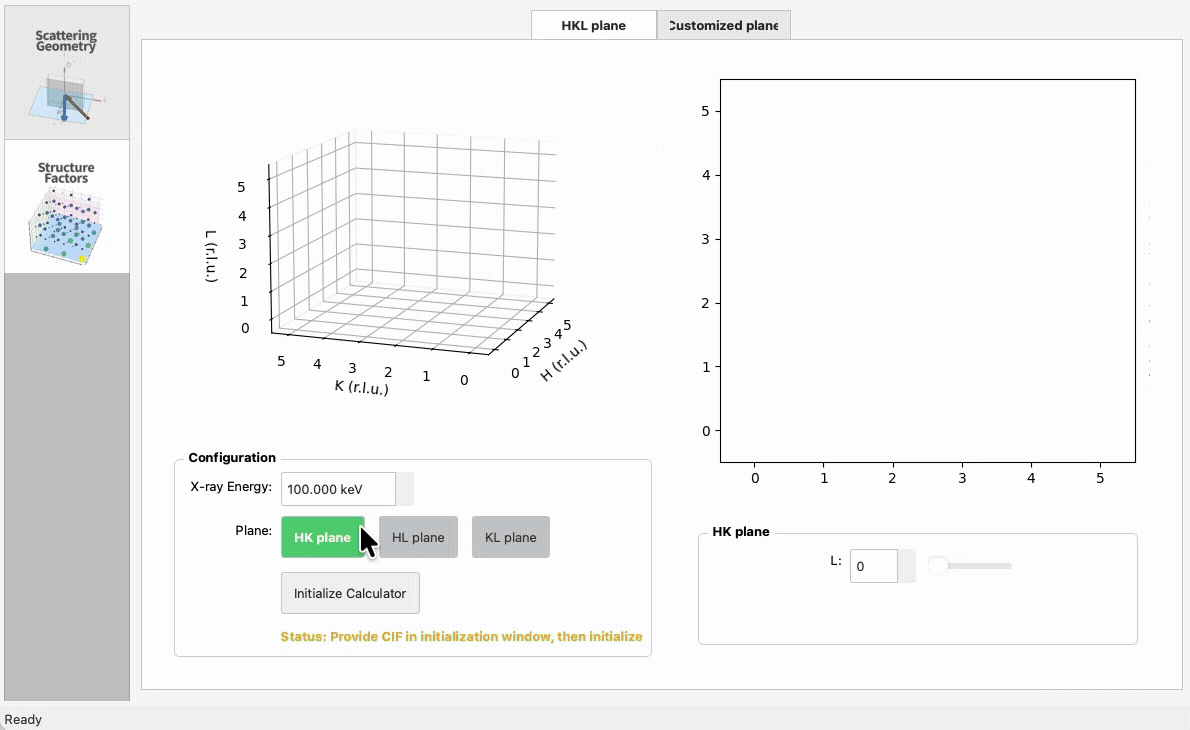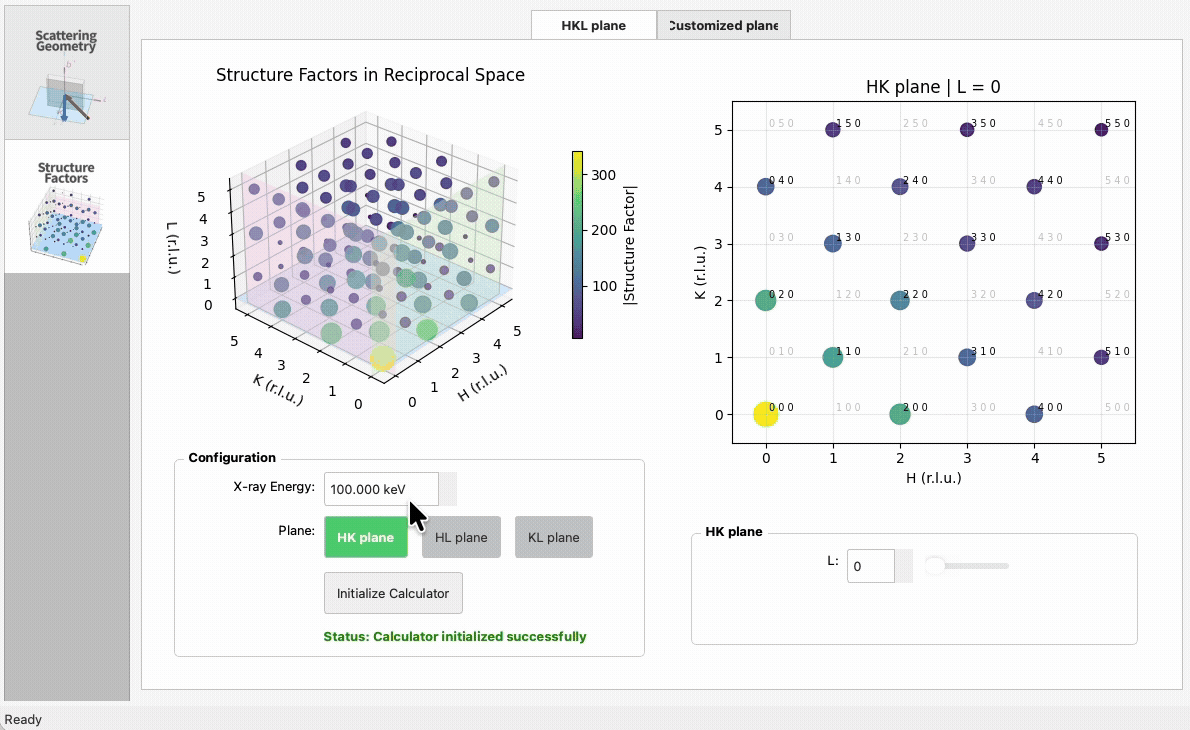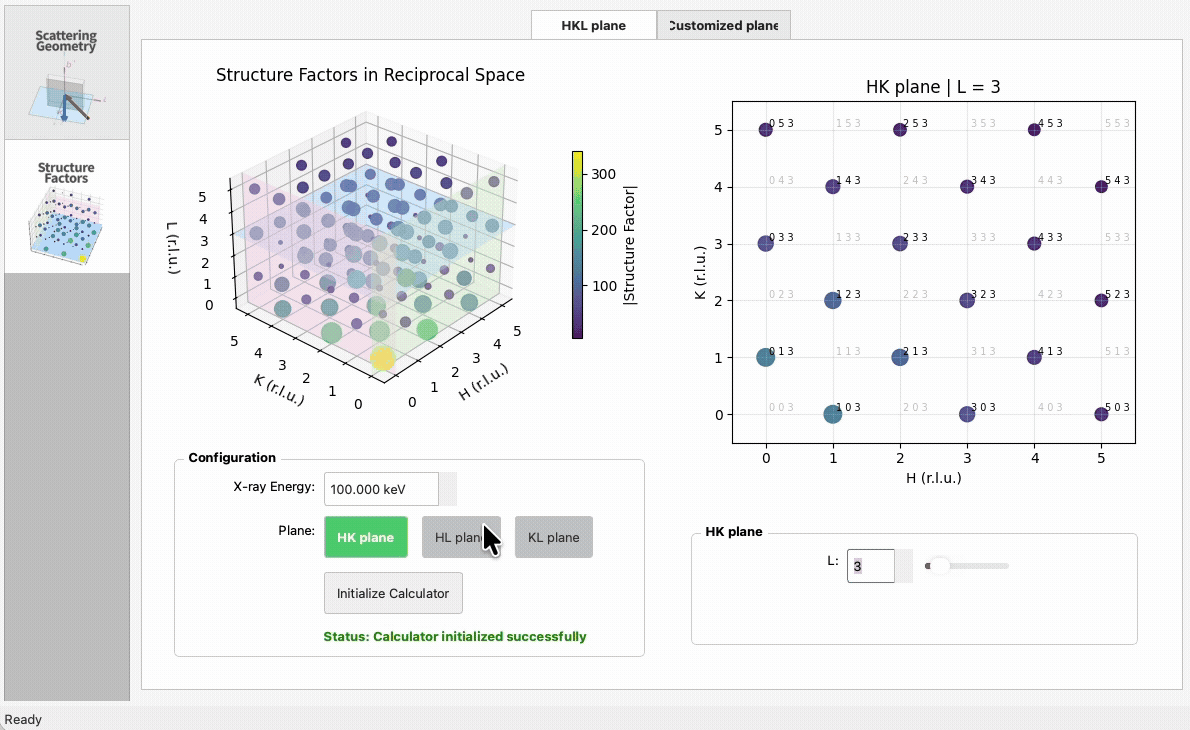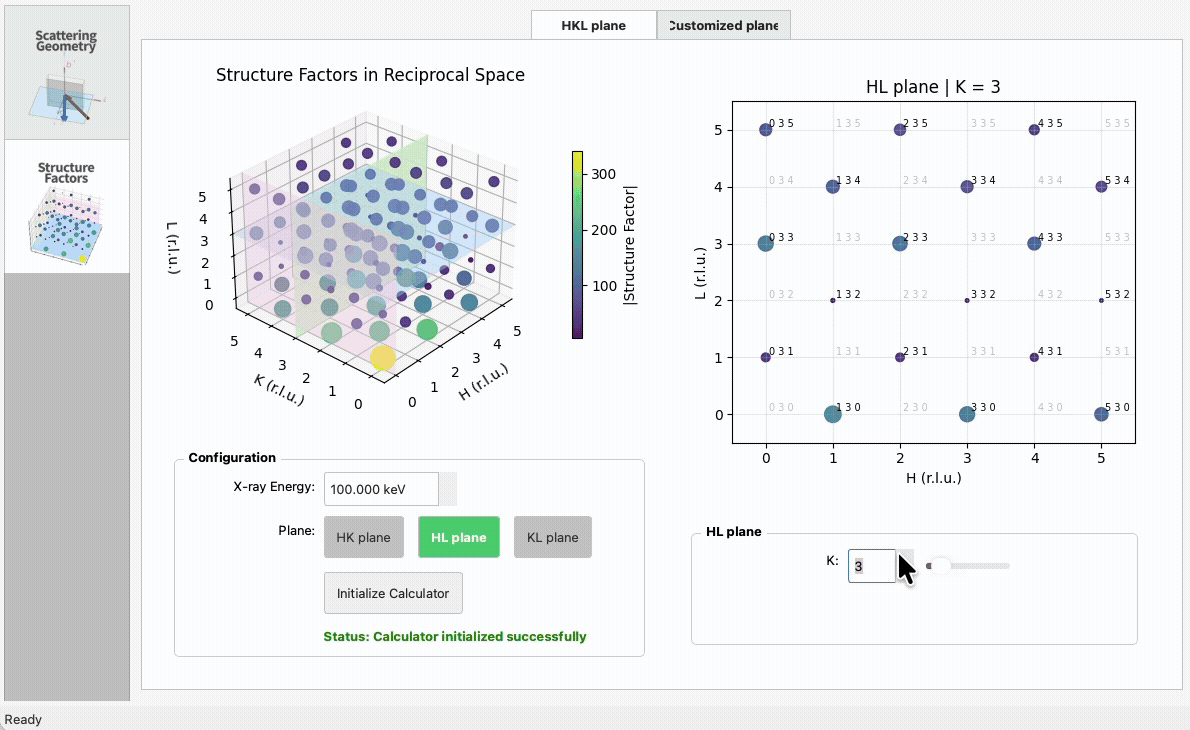
import numpy as np
|
|
5
|
+
from matplotlib.backends.backend_qt5agg import FigureCanvasQTAgg as FigureCanvas
|
|
6
|
+
from matplotlib.figure import Figure
|
|
7
|
+
from mpl_toolkits.mplot3d.art3d import Poly3DCollection
|
|
8
|
+
import Dans_Diffraction as dif
|
|
9
|
+
import matplotlib.cm as cm
|
|
10
|
+
import matplotlib.colors as mcolors
|
|
11
|
+
import matplotlib.pyplot as plt
|
|
12
|
+
|
|
13
|
+
# Import Dans_Diffraction internal functions for plotting
|
|
14
|
+
from Dans_Diffraction import functions_general as fg
|
|
15
|
+
from Dans_Diffraction import functions_plotting as fp
|
|
16
|
+
from Dans_Diffraction import functions_crystallography as fc
|
|
17
|
+
|
|
18
|
+
from advisor.domain import angle_to_matrix, get_rotation
|
|
19
|
+
|
|
20
|
+
# Apply the colormap patch for Dans_Diffraction compatibility
|
|
21
|
+
_old_ensure_cmap = cm._ensure_cmap
|
|
22
|
+
|
|
23
|
+
def _ensure_cmap_patched(cmap):
|
|
24
|
+
if isinstance(cmap, np.ndarray):
|
|
25
|
+
return mcolors.ListedColormap(cmap)
|
|
26
|
+
return _old_ensure_cmap(cmap)
|
|
27
|
+
|
|
28
|
+
cm._ensure_cmap = _ensure_cmap_patched
|
|
29
|
+
|
|
30
|
+
|
|
31
|
+
class UnitcellVisualizer(FigureCanvas):
|
|
32
|
+
def __init__(self, width=4, height=4, dpi=100):
|
|
33
|
+
self.fig = Figure(figsize=(width, height), dpi=dpi)
|
|
34
|
+
self.axes = self.fig.add_subplot(111, projection='3d')
|
|
35
|
+
self.cif_file_path = None
|
|
36
|
+
self._is_initialized = False
|
|
37
|
+
self.crystal = None
|
|
38
|
+
super().__init__(self.fig)
|
|
39
|
+
|
|
40
|
+
# Set background color to white
|
|
41
|
+
self.fig.patch.set_facecolor("white")
|
|
42
|
+
self.axes.set_facecolor("white")
|
|
43
|
+
|
|
44
|
+
# Set initial view
|
|
45
|
+
self.axes.view_init(elev=39, azim=75)
|
|
46
|
+
|
|
47
|
+
# Clean axes like Dans_Diffraction
|
|
48
|
+
self.axes.set_axis_off()
|
|
49
|
+
|
|
50
|
+
@property
|
|
51
|
+
def cif_file_path(self):
|
|
52
|
+
return self._cif_file_path
|
|
53
|
+
|
|
54
|
+
@cif_file_path.setter
|
|
55
|
+
def cif_file_path(self, cif_file_path: str):
|
|
56
|
+
# check if the file exists (allow None for initialization)
|
|
57
|
+
if cif_file_path is not None and not os.path.exists(cif_file_path):
|
|
58
|
+
raise FileNotFoundError(f"File {cif_file_path} does not exist.")
|
|
59
|
+
self._cif_file_path = cif_file_path
|
|
60
|
+
|
|
61
|
+
@property
|
|
62
|
+
def is_initialized(self):
|
|
63
|
+
return self._is_initialized
|
|
64
|
+
|
|
65
|
+
@is_initialized.setter
|
|
66
|
+
def is_initialized(self, is_initialized: bool):
|
|
67
|
+
self._is_initialized = is_initialized
|
|
68
|
+
|
|
69
|
+
|
|
70
|
+
def set_parameters(self, params: dict):
|
|
71
|
+
cif_file_path = params["cif_file"]
|
|
72
|
+
self.cif_file_path = cif_file_path
|
|
73
|
+
try:
|
|
74
|
+
self.crystal = dif.Crystal(self.cif_file_path)
|
|
75
|
+
self.is_initialized = True
|
|
76
|
+
except Exception as e:
|
|
77
|
+
print(f"Error loading crystal structure from {cif_file_path}: {e}")
|
|
78
|
+
self.is_initialized = False
|
|
79
|
+
|
|
80
|
+
def visualize_unitcell(self, show_labels=False, is_clear=True):
|
|
81
|
+
""" read the atom position from the cif file and visualize the unit cell using integrated Dans_Diffraction logic
|
|
82
|
+
"""
|
|
83
|
+
if not self.is_initialized or self.crystal is None:
|
|
84
|
+
print("Unitcell visualizer not initialized.")
|
|
85
|
+
return
|
|
86
|
+
|
|
87
|
+
# Clear the current plot
|
|
88
|
+
self.axes.clear()
|
|
89
|
+
|
|
90
|
+
# Set background color to white and clean axes like Dans_Diffraction
|
|
91
|
+
self.axes.set_facecolor("white")
|
|
92
|
+
self.axes.set_axis_off() # Ensure clean look after clearing
|
|
93
|
+
|
|
94
|
+
try:
|
|
95
|
+
# Direct implementation of Dans_Diffraction plot_crystal logic
|
|
96
|
+
self._plot_crystal_direct(show_labels=show_labels)
|
|
97
|
+
|
|
98
|
+
# Set title with crystal info
|
|
99
|
+
#title = f"Unit Cell: {os.path.basename(self.cif_file_path)}"
|
|
100
|
+
#if hasattr(self.crystal, 'name') and self.crystal.name:
|
|
101
|
+
# title = f"Unit Cell: {self.crystal.name}"
|
|
102
|
+
#title = f"Unit Cell"
|
|
103
|
+
#self.axes.set_title(title, fontsize=12, pad=20)
|
|
104
|
+
|
|
105
|
+
except Exception as e:
|
|
106
|
+
print(f"Error visualizing unit cell: {e}")
|
|
107
|
+
# Fallback: show error message on plot
|
|
108
|
+
self.axes.text(0.5, 0.5, 0.5, f"Error: {str(e)}",
|
|
109
|
+
transform=self.axes.transAxes,
|
|
110
|
+
ha='center', va='center')
|
|
111
|
+
|
|
112
|
+
# Set initial view
|
|
113
|
+
self.axes.view_init(elev=30, azim=55)
|
|
114
|
+
|
|
115
|
+
# Refresh the canvas
|
|
116
|
+
self.draw()
|
|
117
|
+
|
|
118
|
+
def _plot_crystal_direct(self, show_labels=False):
|
|
119
|
+
"""
|
|
120
|
+
Plot crystal structure by rendering atomic positions and unit cell boundaries.
|
|
121
|
+
Based on Dans_Diffraction.classes_plotting.Plotting.plot_crystal method.
|
|
122
|
+
|
|
123
|
+
Args:
|
|
124
|
+
show_labels: If True, display atom labels next to their positions
|
|
125
|
+
"""
|
|
126
|
+
# Tolerance for determining if atoms are within unit cell boundaries
|
|
127
|
+
boundary_tolerance = 0.05
|
|
128
|
+
|
|
129
|
+
# Generate atomic positions in fractional coordinates for one unit cell
|
|
130
|
+
fractional_coordinates, element_symbols, atom_labels, occupancies, _, _ = \
|
|
131
|
+
self.crystal.Structure.generate_lattice(1, 1, 1)
|
|
132
|
+
|
|
133
|
+
# Identify unique atom types and create mapping for colors and sizes
|
|
134
|
+
unique_labels, unique_indices, inverse_indices = np.unique(
|
|
135
|
+
atom_labels, return_index=True, return_inverse=True
|
|
136
|
+
)
|
|
137
|
+
unique_element_types = element_symbols[unique_indices]
|
|
138
|
+
|
|
139
|
+
# Assign colors to each unique atom type using rainbow colormap
|
|
140
|
+
atom_colors = plt.cm.rainbow(np.linspace(0, 1, len(unique_element_types)))
|
|
141
|
+
|
|
142
|
+
# Get atomic radii for each element type for visualization
|
|
143
|
+
atom_radii = fc.atom_properties(unique_element_types, 'Radii')
|
|
144
|
+
|
|
145
|
+
# Convert fractional coordinates to Cartesian coordinates
|
|
146
|
+
cartesian_positions = self.crystal.Cell.calculateR(fractional_coordinates)
|
|
147
|
+
|
|
148
|
+
# Center the structure at origin for better visualization
|
|
149
|
+
center_position = np.mean(cartesian_positions, axis=0)
|
|
150
|
+
cartesian_positions = cartesian_positions - center_position
|
|
151
|
+
|
|
152
|
+
# Create mask for atoms that are visible: within unit cell bounds and sufficiently occupied
|
|
153
|
+
visible_atom_mask = np.all(
|
|
154
|
+
np.hstack([
|
|
155
|
+
fractional_coordinates < (1 + boundary_tolerance),
|
|
156
|
+
fractional_coordinates > (0 - boundary_tolerance),
|
|
157
|
+
occupancies.reshape([-1, 1]) > 0.2
|
|
158
|
+
]),
|
|
159
|
+
axis=1
|
|
160
|
+
)
|
|
161
|
+
|
|
162
|
+
# Get maximum lattice parameter for setting axis limits and arrow positioning
|
|
163
|
+
lim = np.max(self.crystal.Cell.lp()[:3])
|
|
164
|
+
|
|
165
|
+
# Plot atoms grouped by element type
|
|
166
|
+
for atom_type_index in range(len(unique_element_types)):
|
|
167
|
+
# Calculate total occupancy for this atom type
|
|
168
|
+
total_occupancy = np.array([
|
|
169
|
+
occupancies[atom_idx]
|
|
170
|
+
for atom_idx in range(len(cartesian_positions))
|
|
171
|
+
if inverse_indices[atom_idx] == atom_type_index
|
|
172
|
+
])
|
|
173
|
+
|
|
174
|
+
# Skip atom types with zero occupancy
|
|
175
|
+
if sum(total_occupancy) == 0:
|
|
176
|
+
continue
|
|
177
|
+
|
|
178
|
+
# Get positions for atoms of this type
|
|
179
|
+
atom_positions = np.array([
|
|
180
|
+
cartesian_positions[atom_idx, :]
|
|
181
|
+
for atom_idx in range(len(cartesian_positions))
|
|
182
|
+
if inverse_indices[atom_idx] == atom_type_index
|
|
183
|
+
])
|
|
184
|
+
|
|
185
|
+
# Filter to only visible atoms (within unit cell and occupied)
|
|
186
|
+
visible_atoms_mask = np.array([
|
|
187
|
+
visible_atom_mask[atom_idx]
|
|
188
|
+
for atom_idx in range(len(cartesian_positions))
|
|
189
|
+
if inverse_indices[atom_idx] == atom_type_index
|
|
190
|
+
])
|
|
191
|
+
|
|
192
|
+
# Create color array for all atoms of this type
|
|
193
|
+
atom_colors_array = np.tile(
|
|
194
|
+
atom_colors[atom_type_index],
|
|
195
|
+
(len(atom_positions[visible_atoms_mask, :]), 1)
|
|
196
|
+
)
|
|
197
|
+
|
|
198
|
+
# Render atoms as 3D scatter points
|
|
199
|
+
self.axes.scatter(
|
|
200
|
+
atom_positions[visible_atoms_mask, 0],
|
|
201
|
+
atom_positions[visible_atoms_mask, 1],
|
|
202
|
+
atom_positions[visible_atoms_mask, 2],
|
|
203
|
+
s=1 * atom_radii[atom_type_index],
|
|
204
|
+
c=atom_colors_array,
|
|
205
|
+
label=unique_labels[atom_type_index],
|
|
206
|
+
alpha=1,
|
|
207
|
+
edgecolors='white',
|
|
208
|
+
linewidth=0.1
|
|
209
|
+
)
|
|
210
|
+
|
|
211
|
+
# Optionally display atom labels
|
|
212
|
+
if show_labels:
|
|
213
|
+
structure_fractional_coords, _, structure_atom_labels, _, _, _ = \
|
|
214
|
+
self.crystal.Structure.get()
|
|
215
|
+
structure_cartesian_positions = self.crystal.Cell.calculateR(
|
|
216
|
+
structure_fractional_coords
|
|
217
|
+
) - center_position
|
|
218
|
+
|
|
219
|
+
for atom_index in range(len(structure_cartesian_positions)):
|
|
220
|
+
self.axes.text(
|
|
221
|
+
structure_cartesian_positions[atom_index, 0],
|
|
222
|
+
structure_cartesian_positions[atom_index, 1],
|
|
223
|
+
structure_cartesian_positions[atom_index, 2],
|
|
224
|
+
'%2d: %s' % (atom_index, structure_atom_labels[atom_index]),
|
|
225
|
+
fontsize=8,
|
|
226
|
+
bbox=dict(boxstyle="round,pad=0.2", facecolor='white', alpha=0.8)
|
|
227
|
+
)
|
|
228
|
+
|
|
229
|
+
# Define unit cell box vertices in fractional coordinates
|
|
230
|
+
unit_cell_box_coords = np.array([
|
|
231
|
+
[0., 0, 0], [1, 0, 0], [1, 0, 1], [1, 1, 1], [1, 1, 0], [0, 1, 0], [0, 1, 1],
|
|
232
|
+
[0, 0, 1], [1, 0, 1], [1, 0, 0], [1, 1, 0], [1, 1, 1], [0, 1, 1], [0, 1, 0],
|
|
233
|
+
[0, 0, 0], [0, 0, 1]
|
|
234
|
+
])
|
|
235
|
+
|
|
236
|
+
# Convert box coordinates to Cartesian and center
|
|
237
|
+
bpos = self.crystal.Cell.calculateR(unit_cell_box_coords) - center_position
|
|
238
|
+
|
|
239
|
+
# Draw unit cell boundary as gray wireframe
|
|
240
|
+
self.axes.plot(bpos[:, 0], bpos[:, 1], bpos[:, 2], c='gray', linewidth=1, alpha=0.85)
|
|
241
|
+
|
|
242
|
+
# Position coordinate system arrows in corner away from structure
|
|
243
|
+
arrow_origin = np.array([-lim * 0.4, -lim * 0.4, -lim * 0.4])
|
|
244
|
+
arrow_scale = lim * 0.15
|
|
245
|
+
|
|
246
|
+
# Get normalized lattice vectors from the crystal
|
|
247
|
+
a_vec = np.array([1, 0, 0])
|
|
248
|
+
b_vec = np.array([0, 1, 0])
|
|
249
|
+
c_vec = np.array([0, 0, 1])
|
|
250
|
+
|
|
251
|
+
# Transform to real space coordinates
|
|
252
|
+
a_real = self.crystal.Cell.calculateR(a_vec.reshape(1, -1))[0]
|
|
253
|
+
b_real = self.crystal.Cell.calculateR(b_vec.reshape(1, -1))[0]
|
|
254
|
+
c_real = self.crystal.Cell.calculateR(c_vec.reshape(1, -1))[0]
|
|
255
|
+
|
|
256
|
+
# Normalize and scale the vectors
|
|
257
|
+
a_norm = a_real / np.linalg.norm(a_real) * arrow_scale
|
|
258
|
+
b_norm = b_real / np.linalg.norm(b_real) * arrow_scale
|
|
259
|
+
c_norm = c_real / np.linalg.norm(c_real) * arrow_scale
|
|
260
|
+
|
|
261
|
+
# Draw coordinate arrows with labels
|
|
262
|
+
self.axes.quiver(arrow_origin[0], arrow_origin[1], arrow_origin[2]+ 1.6*bpos[7, 2],
|
|
263
|
+
a_norm[0], a_norm[1], a_norm[2],
|
|
264
|
+
color='dodgerblue', arrow_length_ratio=0.15, linewidth=2, alpha=0.9)
|
|
265
|
+
self.axes.text(arrow_origin[0] + a_norm[0]*1.2, arrow_origin[1] + a_norm[1]*1,
|
|
266
|
+
arrow_origin[2] + a_norm[2]*1.2 + 1.6*bpos[7, 2], 'a', color='dodgerblue', fontsize=14, fontweight='bold')
|
|
267
|
+
|
|
268
|
+
self.axes.quiver(arrow_origin[0], arrow_origin[1], arrow_origin[2]+ 1.6*bpos[7, 2],
|
|
269
|
+
b_norm[0], b_norm[1], b_norm[2],
|
|
270
|
+
color='dodgerblue', arrow_length_ratio=0.15, linewidth=2, alpha=0.9)
|
|
271
|
+
self.axes.text(arrow_origin[0] + b_norm[0]*1.2, arrow_origin[1] + b_norm[1]*1.2,
|
|
272
|
+
arrow_origin[2] + b_norm[2]*1.2 + 1.6*bpos[7, 2], 'b', color='dodgerblue', fontsize=14, fontweight='bold')
|
|
273
|
+
|
|
274
|
+
self.axes.quiver(arrow_origin[0], arrow_origin[1], arrow_origin[2]+ 1.6*bpos[7, 2],
|
|
275
|
+
c_norm[0], c_norm[1], c_norm[2],
|
|
276
|
+
color='dodgerblue', arrow_length_ratio=0.15, linewidth=2, alpha=0.9)
|
|
277
|
+
self.axes.text(arrow_origin[0] + c_norm[0]*1.2, arrow_origin[1] + c_norm[1]*1.2,
|
|
278
|
+
arrow_origin[2] + c_norm[2]*1.2 + 1.6*bpos[7, 2], 'c', color='dodgerblue', fontsize=14, fontweight='bold')
|
|
279
|
+
|
|
280
|
+
# Set axis limits based on lattice
|
|
281
|
+
self.axes.set_xlim(-lim/2, lim/2)
|
|
282
|
+
self.axes.set_ylim(-lim/2, lim/2)
|
|
283
|
+
self.axes.set_zlim(-lim/2, lim/2)
|
|
284
|
+
|
|
285
|
+
# Turn off axis (removes background, grids, ticks, labels) - like Dans_Diffraction
|
|
286
|
+
#self.axes.set_axis_off()
|
|
287
|
+
# change view angles
|
|
288
|
+
# Add legend positioned closer to the unit cell
|
|
289
|
+
self.axes.legend(fontsize=10, frameon=False, loc='upper left',
|
|
290
|
+
bbox_to_anchor=(0.98, 0.98), handletextpad=0.3,
|
|
291
|
+
handlelength=1.5, columnspacing=0.1,
|
|
292
|
+
fancybox=False, shadow=False)
|
|
293
|
+
self.fig.tight_layout()
|
|
294
|
+
|
|
295
|
+
|
|
296
|
+
|
|
297
|
+
|
|
298
|
+
def visualize_scattering_geometry(self, scattering_angles=None, is_clear=False):
|
|
299
|
+
""" plotting the scattering plane and the beam as done in the ScatteringVisualizer
|
|
300
|
+
"""
|
|
301
|
+
if not self.is_initialized or self.crystal is None:
|
|
302
|
+
print("Unitcell visualizer not initialized.")
|
|
303
|
+
return
|
|
304
|
+
if is_clear:
|
|
305
|
+
# Clear previous plot
|
|
306
|
+
self.axes.clear()
|
|
307
|
+
# Plot the x-ray beam
|
|
308
|
+
if scattering_angles is None:
|
|
309
|
+
scattering_angles = {
|
|
310
|
+
"theta": 50,
|
|
311
|
+
"tth": 150,
|
|
312
|
+
"phi": 0,
|
|
313
|
+
"chi": 0,
|
|
314
|
+
}
|
|
315
|
+
tth, theta, phi, chi = scattering_angles.get("tth", 150), scattering_angles.get("theta", 50), scattering_angles.get("phi", 0), scattering_angles.get("chi", 0)
|
|
316
|
+
a_vec = np.array([1, 0, 0])
|
|
317
|
+
b_vec = np.array([0, 1, 0])
|
|
318
|
+
c_vec = np.array([0, 0, 1])
|
|
319
|
+
|
|
320
|
+
# Transform to real space coordinates
|
|
321
|
+
a_real = self.crystal.Cell.calculateR(a_vec.reshape(1, -1))[0]
|
|
322
|
+
b_real = self.crystal.Cell.calculateR(b_vec.reshape(1, -1))[0]
|
|
323
|
+
c_real = self.crystal.Cell.calculateR(c_vec.reshape(1, -1))[0]
|
|
324
|
+
|
|
325
|
+
|
|
326
|
+
|
|
327
|
+
# Get scale factor from unit cell if available
|
|
328
|
+
if self.is_initialized and self.crystal is not None:
|
|
329
|
+
lim = np.max(self.crystal.Cell.lp()[:3])
|
|
330
|
+
scale_factor = lim / 2 * 1.5 # Match the unit cell scale
|
|
331
|
+
else:
|
|
332
|
+
scale_factor = 1.5 # Default scale if no crystal loaded
|
|
333
|
+
# Plot the scattering plane - scale it to match unit cell, adjusted for beam from -y
|
|
334
|
+
plane_width = 0.25 * scale_factor # narrower in x (rotated 90°)
|
|
335
|
+
plane_height_bottom = -0.75 * scale_factor # extends more in y direction
|
|
336
|
+
plane_height_top = 1.25 * scale_factor
|
|
337
|
+
|
|
338
|
+
x_basis = a_real / np.linalg.norm(a_real)
|
|
339
|
+
# y_basis = cross(a,b)
|
|
340
|
+
z_basis = np.cross(a_real, b_real) / np.linalg.norm(np.cross(a_real, b_real))
|
|
341
|
+
y_basis = np.cross(z_basis, x_basis) / np.linalg.norm(np.cross(z_basis, x_basis))
|
|
342
|
+
|
|
343
|
+
# Define vertices in local plane coordinates (x-y plane, z=0)
|
|
344
|
+
local_vertices = np.array([
|
|
345
|
+
[plane_width*4, plane_height_bottom, 0], # bottom right in local coords
|
|
346
|
+
[-plane_width, plane_height_bottom, 0], # bottom left in local coords
|
|
347
|
+
[plane_width*4, plane_height_top, 0], # top right in local coords
|
|
348
|
+
[-plane_width, plane_height_top, 0], # top left in local coords
|
|
349
|
+
])
|
|
350
|
+
|
|
351
|
+
# Transform vertices to real space using crystal basis vectors
|
|
352
|
+
scatter_plane_vertices = np.zeros_like(local_vertices)
|
|
353
|
+
for i, vertex in enumerate(local_vertices):
|
|
354
|
+
# Transform each vertex using the basis vectors
|
|
355
|
+
# x component uses x_basis, y component uses y_basis, z component uses z_basis (normal to plane)
|
|
356
|
+
scatter_plane_vertices[i] = (vertex[0] * x_basis +
|
|
357
|
+
vertex[1] * y_basis +
|
|
358
|
+
vertex[2] * z_basis)
|
|
359
|
+
# coordinate change matrix: change to scattering plane system
|
|
360
|
+
ccm = np.array([x_basis, y_basis, z_basis]).T
|
|
361
|
+
scatter_plane_vertices = _rotate_vertices_wrt_plane(scatter_plane_vertices, ccm, phi, chi)
|
|
362
|
+
scatter_plane_faces = np.array([[0, 1, 3, 2]]) # single face
|
|
363
|
+
self.axes.add_collection3d(
|
|
364
|
+
Poly3DCollection(
|
|
365
|
+
scatter_plane_vertices[scatter_plane_faces],
|
|
366
|
+
facecolors=[0.3510, 0.7850, 0.9330], # light blue
|
|
367
|
+
edgecolors=[0.7, 0.7, 0.7],
|
|
368
|
+
alpha=0.3,
|
|
369
|
+
)
|
|
370
|
+
)
|
|
371
|
+
|
|
372
|
+
|
|
373
|
+
|
|
374
|
+
# Plot incident beam (k_in) - coming from -y direction in local coords
|
|
375
|
+
offset = 0
|
|
376
|
+
k_in_length = 1.3 * scale_factor
|
|
377
|
+
# In local scattering plane coords: -y_basis direction (rotated 90° from -x)
|
|
378
|
+
k_in_vec = - k_in_length * y_basis
|
|
379
|
+
k_in_vec = rotate_vector(k_in_vec, ccm, theta, phi, chi)
|
|
380
|
+
# Draw colored arrow on top
|
|
381
|
+
self.axes.quiver(
|
|
382
|
+
-k_in_vec[0],
|
|
383
|
+
-k_in_vec[1] - offset,
|
|
384
|
+
-k_in_vec[2],
|
|
385
|
+
k_in_vec[0],
|
|
386
|
+
k_in_vec[1],
|
|
387
|
+
k_in_vec[2],
|
|
388
|
+
color=(191 / 255, 44 / 255, 0),
|
|
389
|
+
alpha=1,
|
|
390
|
+
linewidth=5,
|
|
391
|
+
arrow_length_ratio=0.2,
|
|
392
|
+
zorder=10,
|
|
393
|
+
)
|
|
394
|
+
|
|
395
|
+
# Plot scattered beam (k_out) - in x-y plane, rotated 90° to match -y incident beam
|
|
396
|
+
k_out_length = 1.3 * scale_factor
|
|
397
|
+
# Rotated 90° in local coords: incident from -y, scattered at angle tth
|
|
398
|
+
k_out_vec = ccm @ np.array([np.sin(np.radians(tth)), -np.cos(np.radians(tth)), 0]) * k_out_length
|
|
399
|
+
k_out_vec = rotate_vector(k_out_vec, ccm, theta, phi, chi)
|
|
400
|
+
# Draw colored arrow on top
|
|
401
|
+
self.axes.quiver(
|
|
402
|
+
0,
|
|
403
|
+
0 + offset,
|
|
404
|
+
0,
|
|
405
|
+
k_out_vec[0],
|
|
406
|
+
k_out_vec[1],
|
|
407
|
+
k_out_vec[2],
|
|
408
|
+
color=(2 / 255, 78 / 255, 191 / 255),
|
|
409
|
+
linewidth=5,
|
|
410
|
+
arrow_length_ratio=0.2,
|
|
411
|
+
zorder=10,
|
|
412
|
+
)
|
|
413
|
+
|
|
414
|
+
# Set axis limits to match unit cell scale
|
|
415
|
+
if self.is_initialized and self.crystal is not None:
|
|
416
|
+
self.axes.set_xlim(-lim/2, lim/2)
|
|
417
|
+
self.axes.set_ylim(-lim/2, lim/2)
|
|
418
|
+
self.axes.set_zlim(-lim/2, lim/2)
|
|
419
|
+
else:
|
|
420
|
+
# Fallback to default limits
|
|
421
|
+
self.axes.set_xlim(-1, 1)
|
|
422
|
+
self.axes.set_ylim(-1, 1)
|
|
423
|
+
self.axes.set_zlim(-1, 1)
|
|
424
|
+
|
|
425
|
+
#self.axes.set_axis_off()
|
|
426
|
+
# Update the canvas
|
|
427
|
+
self.draw()
|
|
428
|
+
|
|
429
|
+
def clear_plot(self):
|
|
430
|
+
""" clear the plot """
|
|
431
|
+
self.axes.clear()
|
|
432
|
+
self.draw()
|
|
433
|
+
|
|
434
|
+
|
|
435
|
+
def rotate_vector(vec, ccm, theta, phi, chi):
|
|
436
|
+
"""Convert angles theta, phi, chi to rotation matrix. This rotate the beam or the scattering
|
|
437
|
+
plane, not the sample.
|
|
438
|
+
Pay attention to the direction of the rotation.
|
|
439
|
+
|
|
440
|
+
Updated for x-y scattering plane (z-axis is normal to scattering plane).
|
|
441
|
+
|
|
442
|
+
Args:
|
|
443
|
+
vec (np.ndarray): vector to rotate
|
|
444
|
+
ccm (np.ndarray): coordinate change matrix
|
|
445
|
+
theta (float): rotation about the z-axis in degrees, right-hand rule
|
|
446
|
+
phi (float): rotation about the x-axis in degrees, right-hand rule
|
|
447
|
+
chi (float): rotation about the y-axis in degrees, right-hand rule
|
|
448
|
+
|
|
449
|
+
Returns:
|
|
450
|
+
rotated_vec (np.ndarray): Rotated vector
|
|
451
|
+
"""
|
|
452
|
+
|
|
453
|
+
matrix = angle_to_matrix(theta, phi, chi)
|
|
454
|
+
matrix = ccm @ matrix.T @ ccm.T
|
|
455
|
+
return matrix @ vec
|
|
456
|
+
|
|
457
|
+
def _rotate_vertices_wrt_plane(vertices, ccm, phi, chi):
|
|
458
|
+
"""Rotate the vertices of the sample with respect to the scattering plane
|
|
459
|
+
|
|
460
|
+
Updated for x-y scattering plane (z-axis is normal to scattering plane).
|
|
461
|
+
phi: rotation about y-axis
|
|
462
|
+
chi: rotation about x-axis
|
|
463
|
+
"""
|
|
464
|
+
|
|
465
|
+
rotation_matrix = get_rotation(phi, chi)
|
|
466
|
+
rotation_matrix = ccm @ rotation_matrix.T @ ccm.T
|
|
467
|
+
vertices = np.array(vertices)
|
|
468
|
+
for i, vertex in enumerate(vertices):
|
|
469
|
+
vertices[i] = rotation_matrix @ vertex
|
|
470
|
+
return vertices
|
|
471
|
+
|
|
472
|
+
|
|
473
|
+
|
|
474
|
+
|
|
475
|
+
|
|
476
|
+
|
|
477
|
+
if __name__ == "__main__":
|
|
478
|
+
import matplotlib
|
|
479
|
+
import matplotlib.pyplot as plt
|
|
480
|
+
|
|
481
|
+
# Set matplotlib to use a non-interactive backend for testing
|
|
482
|
+
matplotlib.use('Agg') # Use Agg backend for saving files
|
|
483
|
+
|
|
484
|
+
print(f"Using matplotlib backend: {matplotlib.get_backend()}")
|
|
485
|
+
print("Testing UnitcellVisualizer...")
|
|
486
|
+
|
|
487
|
+
# Create visualizer
|
|
488
|
+
viz = UnitcellVisualizer(width=10, height=8)
|
|
489
|
+
|
|
490
|
+
# Test with both available CIF files
|
|
491
|
+
cif_files = ['data/nacl.cif', 'data/LSCO.cif']
|
|
492
|
+
|
|
493
|
+
for cif_file in cif_files:
|
|
494
|
+
if os.path.exists(cif_file):
|
|
495
|
+
print(f"Processing {cif_file}...")
|
|
496
|
+
|
|
497
|
+
# Set CIF file path
|
|
498
|
+
viz.set_parameters({'cif_file': cif_file})
|
|
499
|
+
|
|
500
|
+
# Create visualization
|
|
501
|
+
viz.visualize_unitcell()
|
|
502
|
+
|
|
503
|
+
# Create output filename
|
|
504
|
+
base_name = os.path.splitext(os.path.basename(cif_file))[0]
|
|
505
|
+
output_file = f'figures/unitcell_{base_name}.png'
|
|
506
|
+
|
|
507
|
+
# Ensure figures directory exists
|
|
508
|
+
os.makedirs('figures', exist_ok=True)
|
|
509
|
+
|
|
510
|
+
# Save the plot to file
|
|
511
|
+
viz.fig.savefig(output_file, dpi=150, bbox_inches='tight',
|
|
512
|
+
facecolor='white', edgecolor='none')
|
|
513
|
+
print(f"Saved unit cell visualization to {output_file}")
|
|
514
|
+
|
|
515
|
+
else:
|
|
516
|
+
print(f"CIF file not found: {cif_file}")
|
|
517
|
+
|
|
518
|
+
print("Test completed successfully!")
|
|
@@ -0,0 +1,122 @@
|
|
|
1
|
+
Metadata-Version: 2.4
|
|
2
|
+
Name: advisor-scattering
|
|
3
|
+
Version: 0.5.0
|
|
4
|
+
Summary: Advisor-Scattering: Advanced Visual Scattering Toolkit for Reciprocal-space
|
|
5
|
+
Author: Xunyang Hong
|
|
6
|
+
License: Proprietary
|
|
7
|
+
Project-URL: Homepage, https://github.com/HongXunyang/advisor
|
|
8
|
+
Project-URL: Documentation, https://advisor-scattering.readthedocs.io/en/latest/
|
|
9
|
+
Keywords: scattering,reciprocal-space,pyqt,visualization
|
|
10
|
+
Classifier: Programming Language :: Python :: 3
|
|
11
|
+
Classifier: Programming Language :: Python :: 3 :: Only
|
|
12
|
+
Classifier: License :: Other/Proprietary License
|
|
13
|
+
Classifier: Operating System :: OS Independent
|
|
14
|
+
Classifier: Topic :: Scientific/Engineering
|
|
15
|
+
Requires-Python: >=3.8
|
|
16
|
+
Description-Content-Type: text/markdown
|
|
17
|
+
Requires-Dist: PyQt5>=5.15.11
|
|
18
|
+
Requires-Dist: matplotlib>=3.9.2
|
|
19
|
+
Requires-Dist: numpy>=2.0.2
|
|
20
|
+
Requires-Dist: scipy>=1.13.1
|
|
21
|
+
Requires-Dist: Dans_Diffraction>=3.3.3
|
|
22
|
+
|
|
23
|
+
# Advisor-Scattering — Advanced Visual Scattering Toolkit for Reciprocal-space
|
|
24
|
+
|
|
25
|
+

|
|
26
|
+

|
|
27
|
+
|
|
28
|
+
Advisor-Scattering is a PyQt5 desktop app for X-ray scattering/diffraction experiments. It helps you convert scattering angles ↔ momentum transfer (HKL), explore scattering geometry, and visualize structure factors—all with interactive plots. Full docs on *[Read the Docs](https://advisor-scattering.readthedocs.io/en/latest/)*.
|
|
29
|
+
|
|
30
|
+
|
|
31
|
+
or use the link below to view the demo video.
|
|
32
|
+
▶ [Demo video (MP4)](https://raw.githubusercontent.com/HongXunyang/advisor/main/docs/source/_static/showcase.mp4)
|
|
33
|
+
|
|
34
|
+
|
|
35
|
+
## Features
|
|
36
|
+
- Convert scattering angles to momentum transfer (HKL) and vice versa.
|
|
37
|
+
- Visualize scattering geometry and unit cells
|
|
38
|
+
- Compute and visualize structure factors in reciprocal space.
|
|
39
|
+
- CIF file drop-in support
|
|
40
|
+
|
|
41
|
+
|
|
42
|
+
## Install
|
|
43
|
+
- Python 3.8+ with PyQt5, numpy, scipy, matplotlib, Dans_Diffraction (see `requirements.txt`).
|
|
44
|
+
|
|
45
|
+
From PyPI:
|
|
46
|
+
```bash
|
|
47
|
+
pip install advisor-scattering
|
|
48
|
+
```
|
|
49
|
+
|
|
50
|
+
From source:
|
|
51
|
+
```bash
|
|
52
|
+
python -m venv .venv
|
|
53
|
+
source .venv/bin/activate # .venv\Scripts\activate on Windows
|
|
54
|
+
pip install -r requirements.txt
|
|
55
|
+
pip install .
|
|
56
|
+
```
|
|
57
|
+
|
|
58
|
+
## Run
|
|
59
|
+
```bash
|
|
60
|
+
advisor-scattering
|
|
61
|
+
# or
|
|
62
|
+
advisor
|
|
63
|
+
# or
|
|
64
|
+
python -m advisor
|
|
65
|
+
```
|
|
66
|
+
|
|
67
|
+
*Note: the install command is `pip install advisor-scattering`, and the import is `import advisor`*.
|
|
68
|
+
|
|
69
|
+
------
|
|
70
|
+
|
|
71
|
+
## Minimal workflow (60 seconds)
|
|
72
|
+
1) Launch the app.
|
|
73
|
+
2) Enter lattice constants/angles and beam energy, or drop a CIF file.
|
|
74
|
+
3) Click **Initialize**.
|
|
75
|
+
4) Use the feature tabs (Scattering Geometry / Structure Factor) to calculate and visualize.
|
|
76
|
+
|
|
77
|
+
|
|
78
|
+
|
|
79
|
+
----
|
|
80
|
+
|
|
81
|
+
## Using the app
|
|
82
|
+
|
|
83
|
+
### 1. Initialization window
|
|
84
|
+
- Enter lattice constants (a, b, c) and angles (alpha, beta, gamma); beam energy auto-updates wavelength/|k|.
|
|
85
|
+
- Optional: drop a CIF to autofill lattice parameters and preview the unit cell.
|
|
86
|
+
- Adjust Euler angles (roll, pitch, yaw) to orient the sample relative to the scattering plane;
|
|
87
|
+
- Click **Initialize** to load the main interface and pass parameters to all tabs.
|
|
88
|
+
|
|
89
|
+
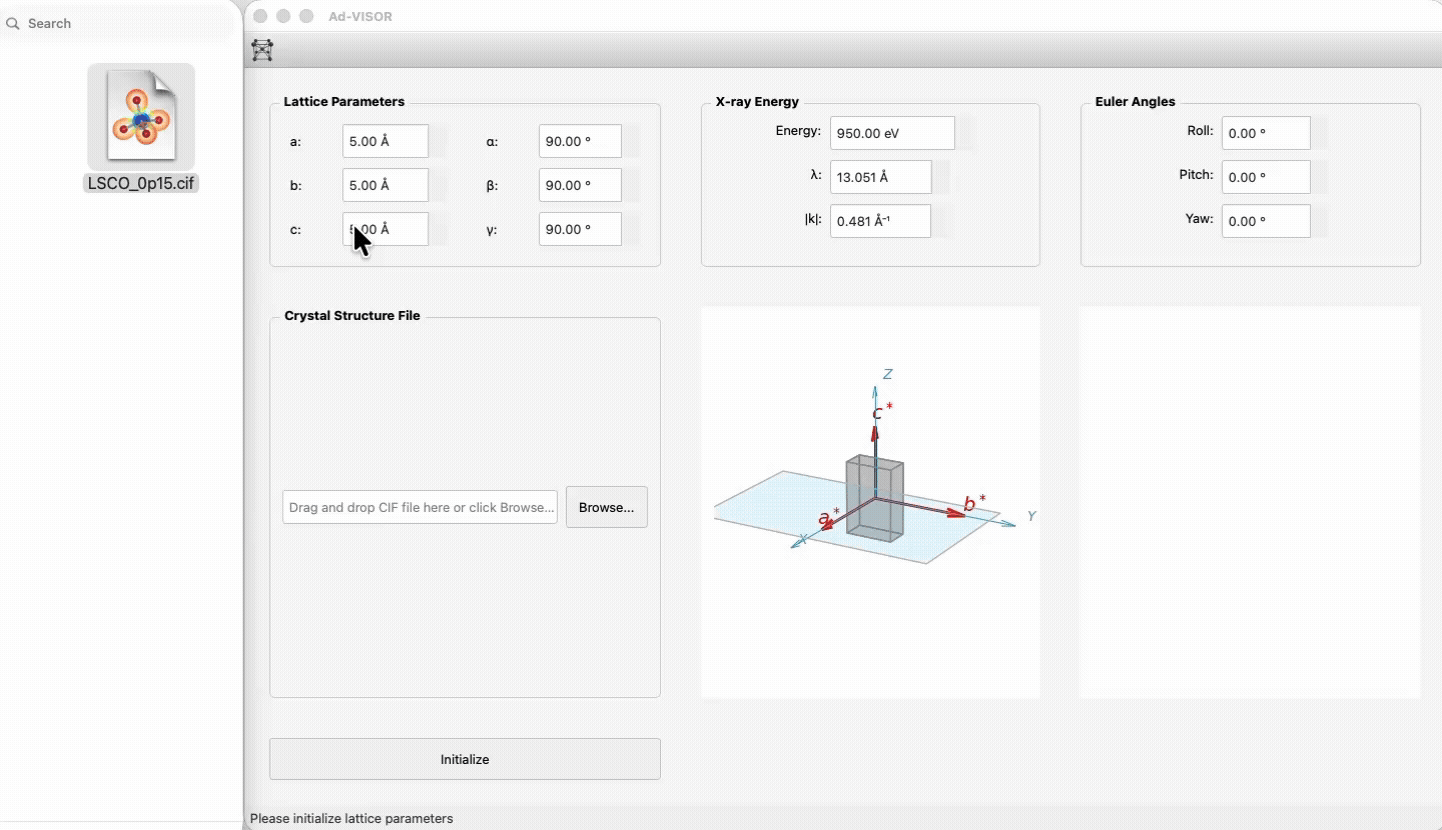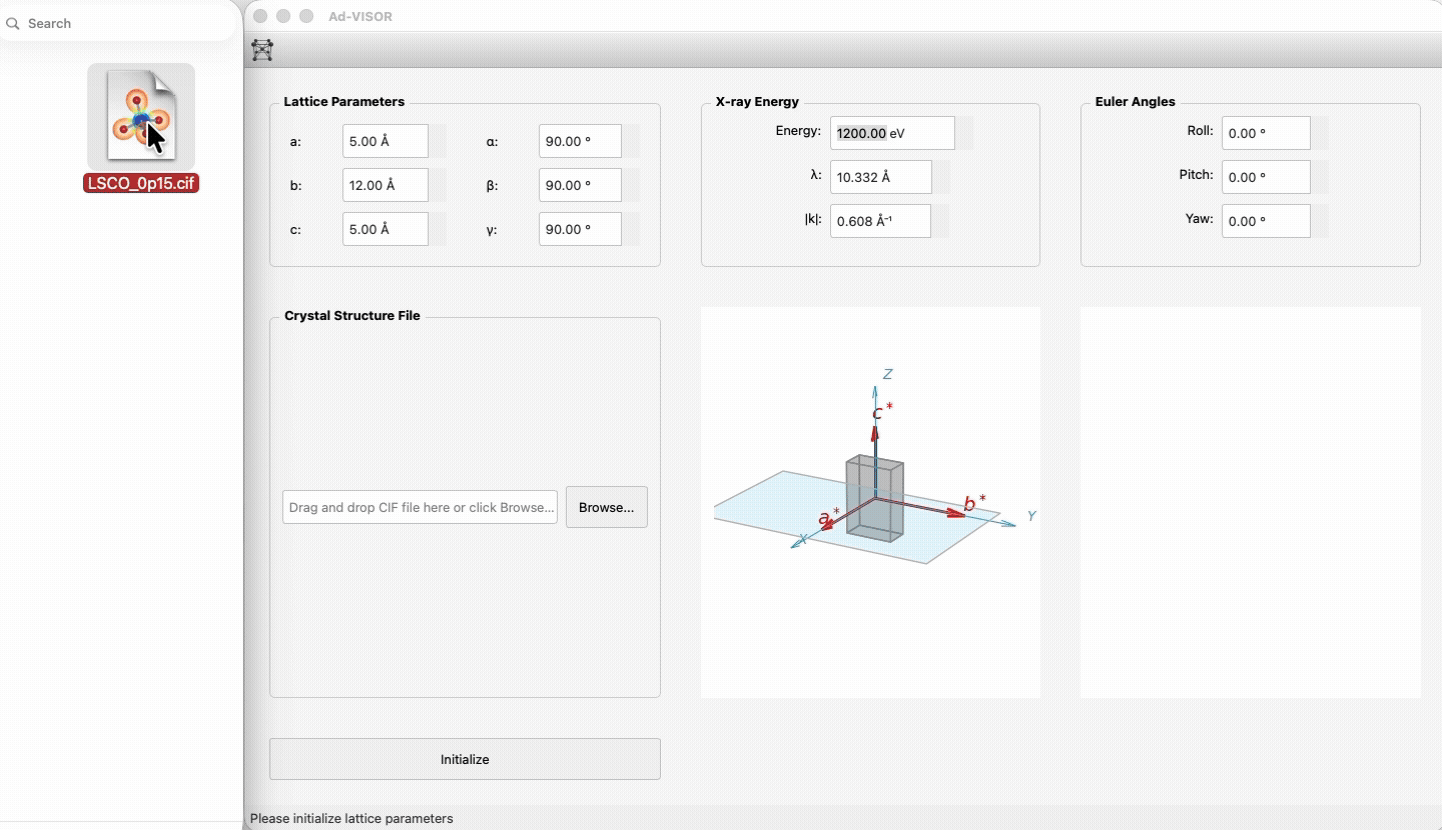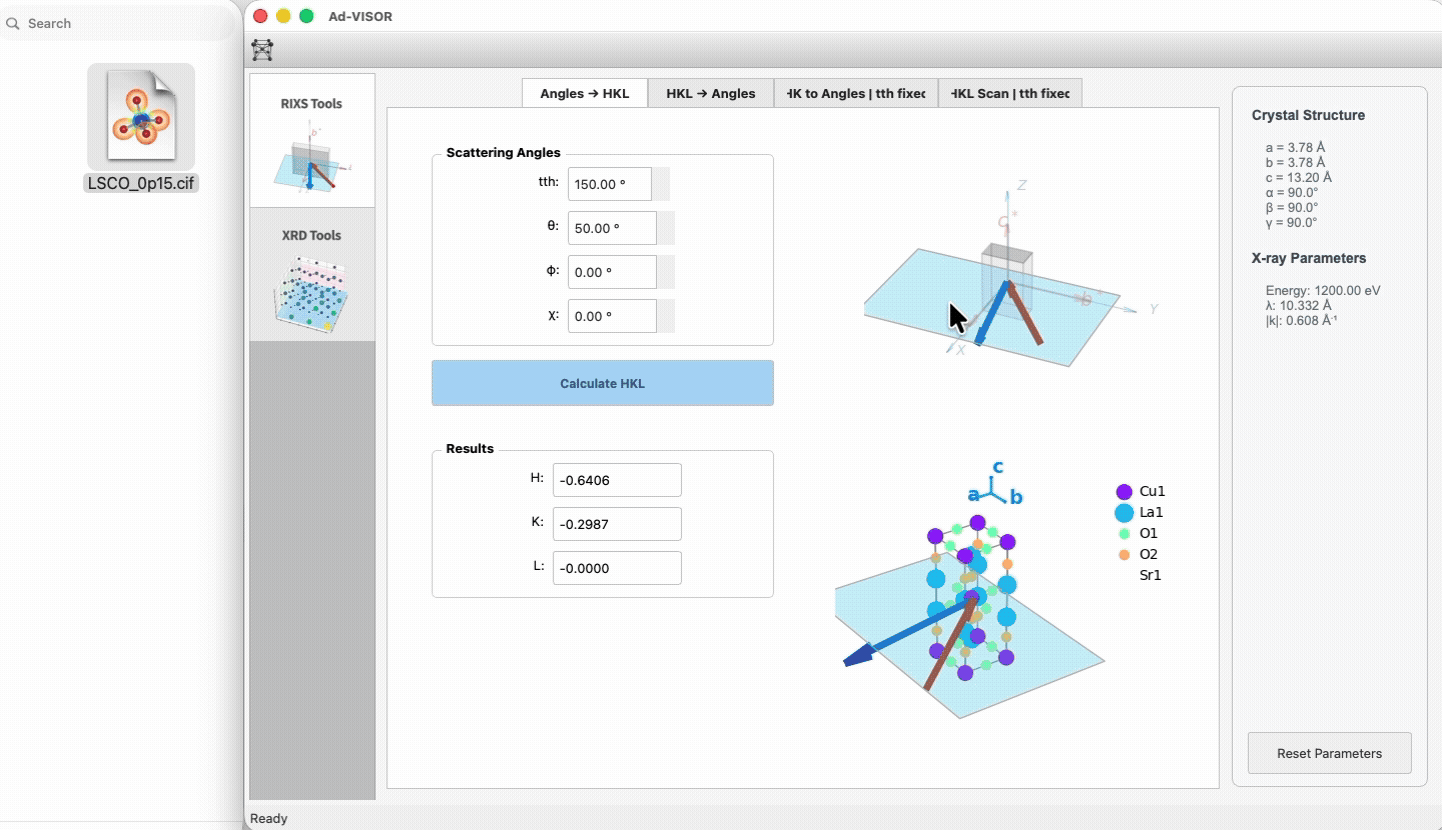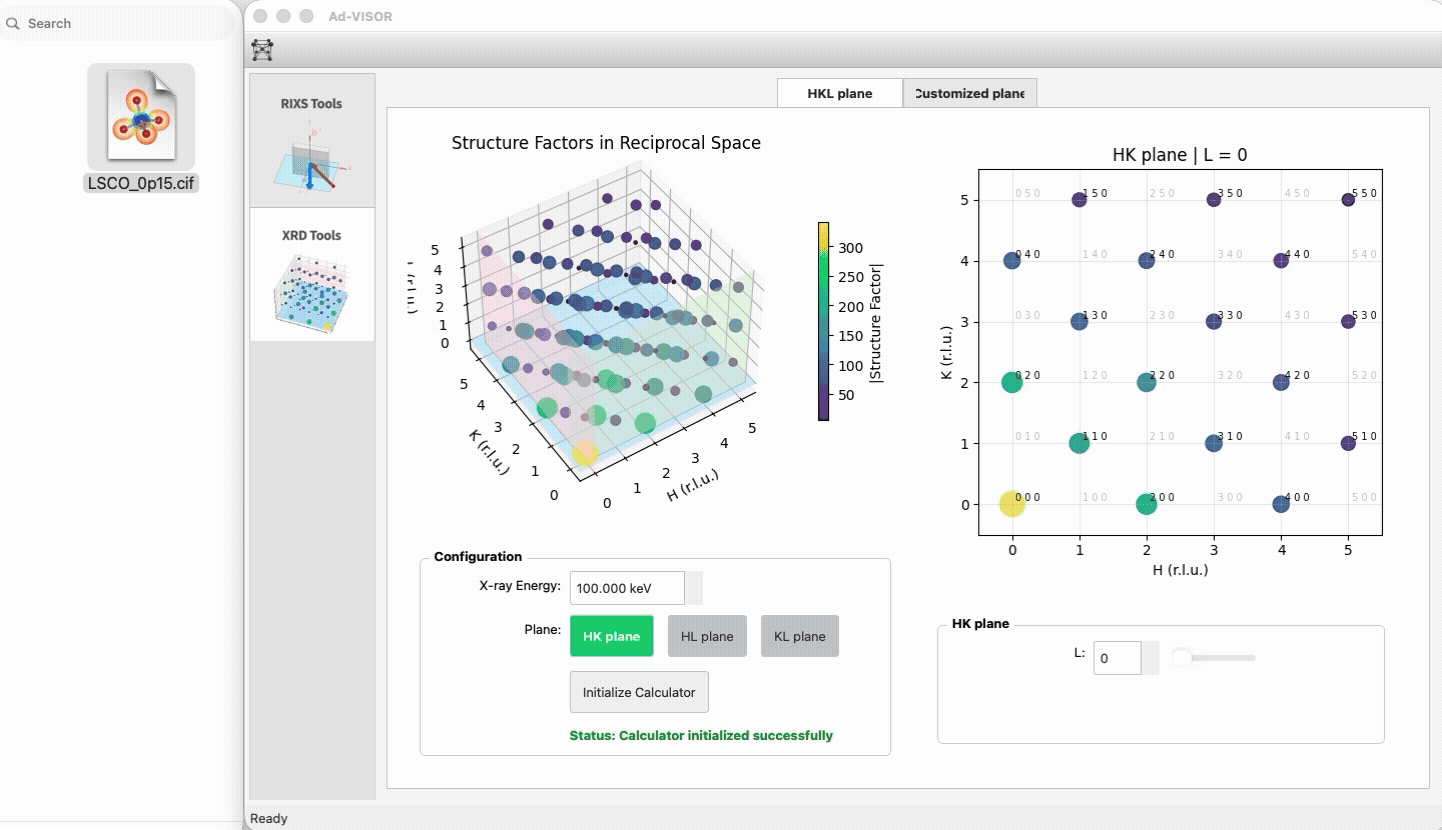
### 2. Scattering Geometry tab
|
|
90
|
+
- Angles → HKL: enter 2θ/θ/χ/φ, compute HKL.
|
|
91
|
+
- HKL → Angles: enter HKL, compute feasible angles.
|
|
92
|
+
- HK to Angles (fixed 2θ) and HKL scan (fixed 2θ) subtabs for trajectory planning.
|
|
93
|
+
|
|
94
|
+
|
|
95
|
+
|
|
96
|
+
### 3. Structure Factor tab
|
|
97
|
+
- Requires a CIF (from init) and an energy in the tab.
|
|
98
|
+
- HKL plane: explore a 3D HKL cube with linked HK/HL/KL slices.
|
|
99
|
+
- Customized plane: choose U/V vectors and a center to sample an arbitrary plane in reciprocal space.
|
|
100
|
+
|
|
101
|
+
|
|
102
|
+
|
|
103
|
+
### 4. Resetting
|
|
104
|
+
Use the toolbar button or **File → Reset Parameters** to return to the init window, clear the CIF lock, and re-enter parameters.
|
|
105
|
+
|
|
106
|
+
------
|
|
107
|
+
|
|
108
|
+
## Project structure (at a glance)
|
|
109
|
+
```
|
|
110
|
+
advisor/ application package
|
|
111
|
+
app.py bootstrap
|
|
112
|
+
controllers/ app/feature coordinators
|
|
113
|
+
domain/ math and geometry helpers (no PyQt)
|
|
114
|
+
features/ per-feature domain/controller/ui code
|
|
115
|
+
ui/ shared UI pieces (init window, main window, base tab, visualizers)
|
|
116
|
+
resources/ QSS, icons, config JSON, sample data
|
|
117
|
+
docs/ Sphinx sources and assets
|
|
118
|
+
```
|
|
119
|
+
|
|
120
|
+
## Documentation
|
|
121
|
+
- Full Sphinx docs live in `docs/` and on *[Read the Docs](https://advisor-scattering.readthedocs.io/en/latest/)*.
|
|
122
|
+
- Appendix covers scattering angle definitions and HKL conventions.
|