@teselagen/ove 0.3.15 → 0.3.16
This diff represents the content of publicly available package versions that have been released to one of the supported registries. The information contained in this diff is provided for informational purposes only and reflects changes between package versions as they appear in their respective public registries.
- package/README.md +718 -0
- package/index.cjs.js +197028 -0
- package/index.es.js +196974 -0
- package/index.js +133 -129
- package/index.mjs +133 -129
- package/index.umd.js +109 -148
- package/package.json +3 -3
package/README.md
ADDED
|
@@ -0,0 +1,718 @@
|
|
|
1
|
+
[](https://dashboard.cypress.io/projects/mp89gp/runs)
|
|
2
|
+
|
|
3
|
+
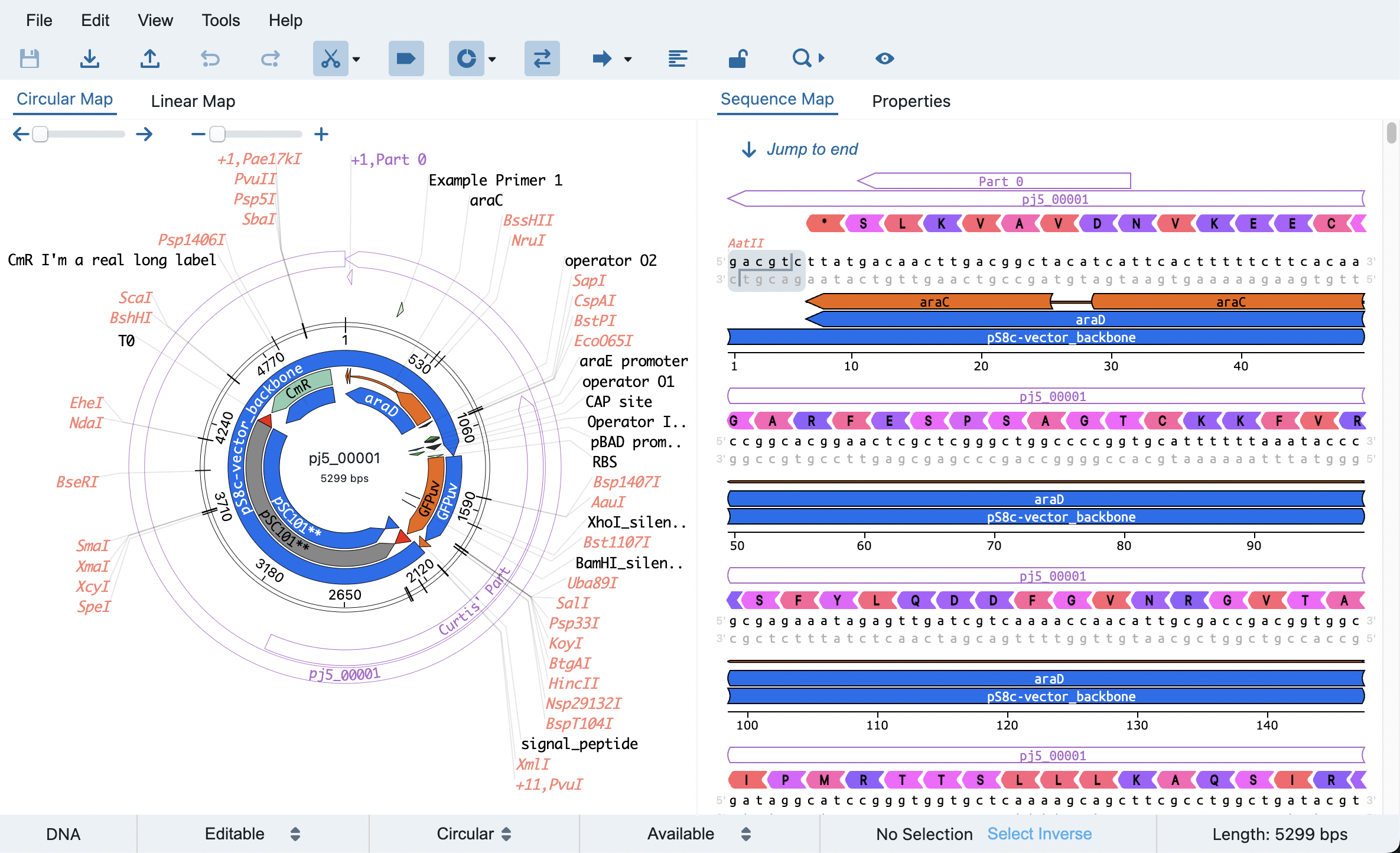
Congrats, you've made it to the repo for Teselagen's Open Source Vector Editor Component
|
|
4
|
+
|
|
5
|
+
- Built With React & Redux
|
|
6
|
+
- Built for easy extensibility + embed-ibility
|
|
7
|
+
- Customize the views to suit your needs
|
|
8
|
+
|
|
9
|
+
|
|
10
|
+
|
|
11
|
+
## [CHANGELOG](CHANGELOG.md)
|
|
12
|
+
|
|
13
|
+
## Demo: https://teselagen.github.io/tg-oss/ove/#/Editor
|
|
14
|
+
|
|
15
|
+
## Get the desktop app: https://github.com/teselagen/ove-electron#installation-instructions
|
|
16
|
+
|
|
17
|
+
## Cypress Tests Dashboard: https://dashboard.cypress.io/#/projects/1zj5vc/runs (you'll need to make a login with cypress)
|
|
18
|
+
|
|
19
|
+
## OVE out in the wild (Who's using OVE):
|
|
20
|
+
|
|
21
|
+
- Teselagen - https://teselagen.com/ (Combinatorial DNA design + Lab management + Ordering + Biohub)
|
|
22
|
+
- ove-electron - https://github.com/teselagen/ove-electron (Simple desktop plasmid editor a la ApE)
|
|
23
|
+
- JBEI - https://bioparts.org/ (Auto annotation and blast tool)
|
|
24
|
+
- JBEI - https://public-registry.jbei.org/ (Lab management and design)
|
|
25
|
+
- Labii - https://docs.labii.com/widgets/biology/@teselagen/ove (Lab management)
|
|
26
|
+
- eLabFTW - https://www.elabftw.net/ (Open source ELN)
|
|
27
|
+
- Colabra - https://www.colabra.ai/ (Lab management, ELN, and R&D AI Copilot) [Docs](https://docs.colabra.ai/guide/reference/run-your-experiments/slash-commands/dna-sequences)
|
|
28
|
+
|
|
29
|
+
# Table of Contents
|
|
30
|
+
|
|
31
|
+
<!-- TOC -->
|
|
32
|
+
|
|
33
|
+
- [[CHANGELOG](CHANGELOG.md)](#changelogchangelogmd)
|
|
34
|
+
- [Demo: https://teselagen.github.io/tg-oss/ove/#/Editor](#demo-httpsteselagengithubiotg-ossoveeditor)
|
|
35
|
+
- [Get the desktop app: https://github.com/teselagen/ove-electron#installation-instructions](#get-the-desktop-app-httpsgithubcomteselagenove-electroninstallation-instructions)
|
|
36
|
+
- [Cypress Tests Dashboard: https://dashboard.cypress.io/#/projects/1zj5vc/runs (you'll need to make a login with cypress)](#cypress-tests-dashboard-httpsdashboardcypressioprojects1zj5vcruns-youll-need-to-make-a-login-with-cypress)
|
|
37
|
+
- [OVE out in the wild (Who's using OVE):](#ove-out-in-the-wild-whos-using-ove)
|
|
38
|
+
- [Table of Contents](#table-of-contents)
|
|
39
|
+
- [Upgrade Instructions for Major and Minor Versions](#upgrade-instructions-for-major-and-minor-versions)
|
|
40
|
+
- [Using this module in React](#using-this-module-in-react)
|
|
41
|
+
- [Example CRA repo with OVE](#example-cra-repo-with-ove)
|
|
42
|
+
- [Installation (react)](#installation-react)
|
|
43
|
+
- [Code (react)](#code-react)
|
|
44
|
+
- [Editor](#editor)
|
|
45
|
+
- [SimpleCircularOrLinearView](#simplecircularorlinearview)
|
|
46
|
+
- [EnzymeViewer](#enzymeviewer)
|
|
47
|
+
- [Using this module outside of react apps (Universal):](#using-this-module-outside-of-react-apps-universal)
|
|
48
|
+
- [Installation (Universal)](#installation-universal)
|
|
49
|
+
- [via npm:](#via-npm)
|
|
50
|
+
- [Or via CDN:](#or-via-cdn)
|
|
51
|
+
- [Full Example:](#full-example)
|
|
52
|
+
- [Code (Universal)](#code-universal)
|
|
53
|
+
- [Accessing the editor state:](#accessing-the-editor-state)
|
|
54
|
+
- [Demo (Universal): http://teselagen.github.io/openVectorEditor/](#demo-universal-httpteselagengithubioopenvectoreditor)
|
|
55
|
+
- [editorProps](#editorprops)
|
|
56
|
+
- [editorState](#editorstate)
|
|
57
|
+
- [Setting Local Storage Persisted Values](#setting-local-storage-persisted-values)
|
|
58
|
+
- [Amino Acid Coloring](#amino-acid-coloring)
|
|
59
|
+
- [Data Model](#data-model)
|
|
60
|
+
- [Feature Locations (aka Feature Joins)](#feature-locations-aka-feature-joins)
|
|
61
|
+
- [Protein Editor](#protein-editor)
|
|
62
|
+
- [Alignments](#alignments)
|
|
63
|
+
- [Integrating your own alignment data (only necessary if not using the built in alignment creation tool)](#integrating-your-own-alignment-data-only-necessary-if-not-using-the-built-in-alignment-creation-tool)
|
|
64
|
+
- [Accessing the alignment state:](#accessing-the-alignment-state)
|
|
65
|
+
- [Alignment Track Data Model](#alignment-track-data-model)
|
|
66
|
+
- [Chromatogram Data](#chromatogram-data)
|
|
67
|
+
- [VersionHistoryView](#versionhistoryview)
|
|
68
|
+
- [Flavors of use (aka Embedded in the Editor vs Standalone and UMD vs React):](#flavors-of-use-aka-embedded-in-the-editor-vs-standalone-and-umd-vs-react)
|
|
69
|
+
- [API:](#api)
|
|
70
|
+
- [getSequenceAtVersion](#getsequenceatversion)
|
|
71
|
+
- [getVersionList](#getversionlist)
|
|
72
|
+
- [onSave [optional] (not necessary unless using the standalone VersionHistoryView)](#onsave-optional-not-necessary-unless-using-the-standalone-versionhistoryview)
|
|
73
|
+
- [exitVersionHistoryView [optional] (not necessary unless using the standalone VersionHistoryView)](#exitversionhistoryview-optional-not-necessary-unless-using-the-standalone-versionhistoryview)
|
|
74
|
+
- [getCurrentSequenceData [optional] (not necessary unless using the standalone VersionHistoryView)](#getcurrentsequencedata-optional-not-necessary-unless-using-the-standalone-versionhistoryview)
|
|
75
|
+
- [Addons](#addons)
|
|
76
|
+
- [Auto annotate addon:](#auto-annotate-addon)
|
|
77
|
+
- [Implementing Autosave functionality](#implementing-autosave-functionality)
|
|
78
|
+
- [Development:](#development)
|
|
79
|
+
- [Prerequisites](#prerequisites)
|
|
80
|
+
- [Outside Developer Set Up Steps](#outside-developer-set-up-steps)
|
|
81
|
+
|
|
82
|
+
<!-- /TOC -->
|
|
83
|
+
|
|
84
|
+
# Upgrade Instructions for Major and Minor Versions
|
|
85
|
+
|
|
86
|
+
This repo follows semantic versioning (major/minor/patche)
|
|
87
|
+
|
|
88
|
+
The commit log can be seen here:
|
|
89
|
+
https://github.com/TeselaGen/openVectorEditor/commits/master
|
|
90
|
+
|
|
91
|
+
Upgrade instructions for any major or minor change can be found here:
|
|
92
|
+
[Upgrade instructions](UPGRADE_INSTRUCTIONS.md)
|
|
93
|
+
|
|
94
|
+
# Using this module in React
|
|
95
|
+
|
|
96
|
+
## Example CRA repo with OVE
|
|
97
|
+
|
|
98
|
+
Here is where you can see how to run the `<Editor/>` in a Create-React-App environment: https://github.com/tnrich/ove-react-demo-repo
|
|
99
|
+
|
|
100
|
+
If you clone that repo you can see it working and see how to set up the redux store/provider and initialize the editor. You should just clone it and run:
|
|
101
|
+
|
|
102
|
+
```
|
|
103
|
+
yarn
|
|
104
|
+
yarn start
|
|
105
|
+
```
|
|
106
|
+
|
|
107
|
+
## Installation (react)
|
|
108
|
+
|
|
109
|
+
```
|
|
110
|
+
yarn add @teselagen/ove
|
|
111
|
+
```
|
|
112
|
+
|
|
113
|
+
## Code (react)
|
|
114
|
+
|
|
115
|
+
Require the following components like:
|
|
116
|
+
|
|
117
|
+
```
|
|
118
|
+
import {Editor, RowView} from "@teselagen/ove"
|
|
119
|
+
```
|
|
120
|
+
|
|
121
|
+
### Editor
|
|
122
|
+
|
|
123
|
+
To use the `<Editor>` component, you'll need to do a bit more work to set up a redux store.
|
|
124
|
+
You can see an example repo where this works here: https://github.com/tnrich/ove-react-demo-repo
|
|
125
|
+
|
|
126
|
+
The `<Editor {...editorProps}/>` component gives you a full blown editor.
|
|
127
|
+
It takes in a list of editorProps as detailed below.
|
|
128
|
+
|
|
129
|
+
### SimpleCircularOrLinearView
|
|
130
|
+
|
|
131
|
+
This is a non-redux connected component that is easy to use:
|
|
132
|
+
|
|
133
|
+
```js
|
|
134
|
+
<SimpleCircularOrLinearView
|
|
135
|
+
sequenceData={{
|
|
136
|
+
circular: true, // or set to false to get a linear view
|
|
137
|
+
sequence: "gagagagag",
|
|
138
|
+
features: [{ id: "lalala", name: "Mr Feature" }]
|
|
139
|
+
}}
|
|
140
|
+
editorName="previewEditor"
|
|
141
|
+
annotationLabelVisibility={{
|
|
142
|
+
//set visibilities as you please
|
|
143
|
+
features: true,
|
|
144
|
+
parts: true,
|
|
145
|
+
cutsites: false,
|
|
146
|
+
primers: true
|
|
147
|
+
}}
|
|
148
|
+
></SimpleCircularOrLinearView>
|
|
149
|
+
```
|
|
150
|
+
|
|
151
|
+
### EnzymeViewer
|
|
152
|
+
|
|
153
|
+
A component used for viewing enzymes
|
|
154
|
+
|
|
155
|
+
```js
|
|
156
|
+
<EnzymeViewer
|
|
157
|
+
{...{
|
|
158
|
+
sequence: "ggtctc",
|
|
159
|
+
reverseSnipPosition: 7,
|
|
160
|
+
forwardSnipPosition: 11
|
|
161
|
+
}}
|
|
162
|
+
/>
|
|
163
|
+
```
|
|
164
|
+
|
|
165
|
+
# Using this module outside of react apps (Universal):
|
|
166
|
+
|
|
167
|
+
The universal build can be used in any app with or without react. It corresponds to using the `<Editor/>` component in the React version. You will be able to customize the Editor just like in the react build, but you will not be able to use the individual components like `<CircularView/>` or `<EnzymeViewer/>`. For that you'll need to use React.
|
|
168
|
+
|
|
169
|
+
## Installation (Universal)
|
|
170
|
+
|
|
171
|
+
### via npm:
|
|
172
|
+
|
|
173
|
+
```
|
|
174
|
+
npm install @teselagen/ove
|
|
175
|
+
```
|
|
176
|
+
|
|
177
|
+
then add the links
|
|
178
|
+
|
|
179
|
+
```html
|
|
180
|
+
<link rel="stylesheet" type="text/css" href="your-path-to-node-modules/@teselagen/ove/umd/style.css" />
|
|
181
|
+
<script type="text/javascript" src="your-path-to-node-modules/@teselagen/ove/index.umd.js.js"></script>
|
|
182
|
+
```
|
|
183
|
+
|
|
184
|
+
### Or via CDN:
|
|
185
|
+
|
|
186
|
+
```html
|
|
187
|
+
<link rel="stylesheet" type="text/css" href="https://unpkg.com/@teselagen/ove/style.css" />
|
|
188
|
+
<script type="text/javascript" src="https://unpkg.com/@teselagen/ove/index.umd.js"></script>
|
|
189
|
+
```
|
|
190
|
+
|
|
191
|
+
### Full Example:
|
|
192
|
+
|
|
193
|
+
A full example of how to set up the unpkg/UMD demo can be seen here: https://github.com/TeselaGen/openVectorEditor/blob/master/demo/public/UMDDemo.html
|
|
194
|
+
Demo here http://teselagen.github.io/openVectorEditor/UMDDemo.html
|
|
195
|
+
|
|
196
|
+
## Code (Universal)
|
|
197
|
+
|
|
198
|
+
```html
|
|
199
|
+
<script>
|
|
200
|
+
const editor = window.createVectorEditor(yourDomNodeHere || "createDomNodeForMe", editorProps); /* createDomNodeForMe will make a dom node for you and append it to the document.body*/
|
|
201
|
+
editor.updateEditor(editorState);
|
|
202
|
+
</script>
|
|
203
|
+
```
|
|
204
|
+
|
|
205
|
+
### Accessing the editor state:
|
|
206
|
+
|
|
207
|
+
```js
|
|
208
|
+
const currentEditorState = editor.getState();
|
|
209
|
+
//you can view various properties of the alignment such as the selection layer using alignment.getState()
|
|
210
|
+
console.info(currentEditorState.selectionLayer);
|
|
211
|
+
```
|
|
212
|
+
|
|
213
|
+
## Demo (Universal): http://teselagen.github.io/openVectorEditor/
|
|
214
|
+
|
|
215
|
+
# editorProps
|
|
216
|
+
|
|
217
|
+
These props consist of hooks and editor config options that can be passed like so: `<Editor {...editorProps}/>` or as seen above like `window.createVectorEditor(yourDomNodeHere, editorProps);`
|
|
218
|
+
|
|
219
|
+
```js
|
|
220
|
+
{
|
|
221
|
+
shouldAutosave: true, //by default the editor does not autosave, setting this to true will trigger the onSave callback after any change to the sequenceData
|
|
222
|
+
//supplying this function WILL make the editor FULLSCREEN BY DEFAULT
|
|
223
|
+
handleFullscreenClose: () => {
|
|
224
|
+
//do whatever you want here
|
|
225
|
+
//UMD only:
|
|
226
|
+
editor.close() //this calls reactDom.unmountComponent at the node you passed as the first arg
|
|
227
|
+
},
|
|
228
|
+
onPreviewModeFullscreenClose: () => {}, //user defined handler for tracking when fullscreen is closed from preview mode
|
|
229
|
+
showReadOnly: false, //default true
|
|
230
|
+
disableSetReadOnly: true, //default false
|
|
231
|
+
onSave: function(event, sequenceDataToSave, editorState, onSuccessCallback) {
|
|
232
|
+
console.info("event:", event);
|
|
233
|
+
console.info("sequenceData:", sequenceDataToSave);
|
|
234
|
+
console.info("editorState:", editorState);
|
|
235
|
+
// To disable the save button after successful saving
|
|
236
|
+
// either call the onSuccessCallback or return a successful promise :)
|
|
237
|
+
onSuccessCallback()
|
|
238
|
+
//or
|
|
239
|
+
// return myPromiseBasedApiCall()
|
|
240
|
+
},
|
|
241
|
+
onCopy: function(event, copiedSequenceData, editorState) {
|
|
242
|
+
//the copiedSequenceData is the subset of the sequence that has been copied in the teselagen sequence format
|
|
243
|
+
console.info("event:", event);
|
|
244
|
+
console.info("sequenceData:", copiedSequenceData);
|
|
245
|
+
console.info("editorState:", editorState);
|
|
246
|
+
const clipboardData = event.clipboardData;
|
|
247
|
+
clipboardData.setData(
|
|
248
|
+
"application/json",
|
|
249
|
+
JSON.stringify(copiedSequenceData)
|
|
250
|
+
);
|
|
251
|
+
event.preventDefault();
|
|
252
|
+
//in onPaste in your app you can do:
|
|
253
|
+
// e.clipboardData.getData('application/json')
|
|
254
|
+
},
|
|
255
|
+
onPaste: function(event, editorState) {
|
|
256
|
+
//the onPaste here must return sequenceData in the teselagen data format
|
|
257
|
+
const clipboardData = event.clipboardData;
|
|
258
|
+
let jsonData = clipboardData.getData("application/json")
|
|
259
|
+
if (jsonData) {
|
|
260
|
+
jsonData = JSON.parse(jsonData)
|
|
261
|
+
if (jsonData.isJbeiSeq) {
|
|
262
|
+
jsonData = convertJbeiToTeselagen(jsonData)
|
|
263
|
+
}
|
|
264
|
+
}
|
|
265
|
+
const sequenceData = jsonData || {sequence: clipboardData.getData("text/plain")}
|
|
266
|
+
return sequenceData
|
|
267
|
+
},
|
|
268
|
+
beforeAnnotationCreate: ({ //also works for edits (!)
|
|
269
|
+
annotationTypePlural, //features/parts/primers
|
|
270
|
+
annotation, //annotation info
|
|
271
|
+
props, //general props to the dialog
|
|
272
|
+
isEdit //true if this is an edit instead of a create
|
|
273
|
+
}) => {
|
|
274
|
+
//a handler to hook into when annotations (features/primers/parts) are being created
|
|
275
|
+
},
|
|
276
|
+
//regular click overrides, eg:
|
|
277
|
+
featureClicked: ({annotation, event}) => {
|
|
278
|
+
//do something here :)
|
|
279
|
+
}
|
|
280
|
+
// orf/primer/translation/cutsite/translationDouble/deletionLayer/replacementLayer/feature/part/searchLayer xxxxClicked can also be overridden
|
|
281
|
+
|
|
282
|
+
rightClickOverrides: { //override what happens when a given feature/part/primer/translation/orf/cutsite/selectionLayer/lineageLine gets right clicked
|
|
283
|
+
//the general format is xxxxRightClicked eg:
|
|
284
|
+
selectionLayerRightClicked: (items, {annotation}, props) => {
|
|
285
|
+
return [...items, {
|
|
286
|
+
//props here get passed directly to blueprintjs MenuItems
|
|
287
|
+
text: "Create Part",
|
|
288
|
+
onClick: () => console.info('hey!≈')
|
|
289
|
+
}]
|
|
290
|
+
}
|
|
291
|
+
},
|
|
292
|
+
PropertiesProps: {
|
|
293
|
+
// the list of tabs shown in the Properties panel
|
|
294
|
+
propertiesList: [
|
|
295
|
+
"general",
|
|
296
|
+
"features",
|
|
297
|
+
"parts",
|
|
298
|
+
"primers",
|
|
299
|
+
"translations",
|
|
300
|
+
"cutsites",
|
|
301
|
+
"orfs",
|
|
302
|
+
"genbank"
|
|
303
|
+
]
|
|
304
|
+
},
|
|
305
|
+
ToolBarProps: {
|
|
306
|
+
toolList: [
|
|
307
|
+
"saveTool",
|
|
308
|
+
//you can override a tool like so:
|
|
309
|
+
{name: "downloadTool", Dropdown: () => {
|
|
310
|
+
return "Hey!"
|
|
311
|
+
}},
|
|
312
|
+
"importTool",
|
|
313
|
+
"undoTool",
|
|
314
|
+
"redoTool",
|
|
315
|
+
"cutsiteTool",
|
|
316
|
+
"featureTool",
|
|
317
|
+
"alignmentTool",
|
|
318
|
+
// "oligoTool",
|
|
319
|
+
"orfTool",
|
|
320
|
+
// "viewTool",
|
|
321
|
+
"editTool",
|
|
322
|
+
"findTool",
|
|
323
|
+
"visibilityTool"
|
|
324
|
+
// "propertiesTool"
|
|
325
|
+
]
|
|
326
|
+
}
|
|
327
|
+
StatusBarProps: {
|
|
328
|
+
//these are the defaults:
|
|
329
|
+
showCircularity: true,
|
|
330
|
+
showReadOnly: true,
|
|
331
|
+
showAvailability: false
|
|
332
|
+
},
|
|
333
|
+
onDigestSave: () => {} //tnr: NOT YET IMPLEMENTED
|
|
334
|
+
}
|
|
335
|
+
```
|
|
336
|
+
|
|
337
|
+
# editorState
|
|
338
|
+
|
|
339
|
+
These are the options to the `updateEditor()` action (the most generic redux action we have)) and will cause the editor state stored in redux to be updated. Only the subset of options that are passed will be updated.
|
|
340
|
+
|
|
341
|
+
```js
|
|
342
|
+
{
|
|
343
|
+
|
|
344
|
+
//note, sequence data passed here will be coerced to fit the Teselagen data model (Teselagen JSON)
|
|
345
|
+
sequenceData: { //Open Vector Editor data model -- https://github.com/TeselaGen/ve-sequence-parsers#format-specification
|
|
346
|
+
sequence: "atagatagagaggcccg",
|
|
347
|
+
features: [
|
|
348
|
+
{
|
|
349
|
+
color: "#b3b3b3", //you can override the default color for each individual feature if you want
|
|
350
|
+
type: "misc_feature",
|
|
351
|
+
start: 0, //start and end are 0-based inclusive for all annotations
|
|
352
|
+
end: 10,
|
|
353
|
+
id: 'yourUniqueID',
|
|
354
|
+
forward: true //ie true=positive strand false=negative strange
|
|
355
|
+
}
|
|
356
|
+
],
|
|
357
|
+
parts: []
|
|
358
|
+
},
|
|
359
|
+
sequenceDataHistory: {}, //clear the sequenceDataHistory if there is any left over from a previous sequence
|
|
360
|
+
sequenceDataHistory: {}, //clear the sequenceDataHistory if there is any left over from a previous sequence
|
|
361
|
+
annotationVisibility: {
|
|
362
|
+
features: false
|
|
363
|
+
},
|
|
364
|
+
panelsShown: [
|
|
365
|
+
[
|
|
366
|
+
{
|
|
367
|
+
id: "sequence",
|
|
368
|
+
name: "Sequence Map",
|
|
369
|
+
active: true
|
|
370
|
+
}
|
|
371
|
+
],
|
|
372
|
+
[
|
|
373
|
+
{
|
|
374
|
+
id: "circular",
|
|
375
|
+
name: "Circular Map",
|
|
376
|
+
active: true
|
|
377
|
+
},
|
|
378
|
+
{
|
|
379
|
+
id: "rail",
|
|
380
|
+
name: "Linear Map",
|
|
381
|
+
active: false
|
|
382
|
+
},
|
|
383
|
+
{
|
|
384
|
+
id: "properties",
|
|
385
|
+
name: "Properties",
|
|
386
|
+
active: false
|
|
387
|
+
}
|
|
388
|
+
]
|
|
389
|
+
],
|
|
390
|
+
caretPosition: 10,
|
|
391
|
+
...additional editor props can be passed here [Example Editor State](./editorStateExample.js)
|
|
392
|
+
}
|
|
393
|
+
```
|
|
394
|
+
|
|
395
|
+
# Setting Local Storage Persisted Values
|
|
396
|
+
|
|
397
|
+
### Amino Acid Coloring
|
|
398
|
+
|
|
399
|
+
```js
|
|
400
|
+
localStorage.setItem("aaColorType", localStorage.getItem("aaColorType") || "byFamily");
|
|
401
|
+
```
|
|
402
|
+
|
|
403
|
+
# Data Model
|
|
404
|
+
|
|
405
|
+
The data model can be interactively inspected by installing the redux devtools for your browser: [devtools](https://chrome.google.com/webstore/detail/redux-devtools/lmhkpmbekcpmknklioeibfkpmmfibljd?hl=en)
|
|
406
|
+
Here is the top level editor state:
|
|
407
|
+
[Example Editor State](./editorStateExample.js)
|
|
408
|
+
|
|
409
|
+
## Feature Locations (aka Feature Joins)
|
|
410
|
+
|
|
411
|
+
Features can have multiple internal locations. You can see an example of a joined feature in the ./editorStateExample.js file linked above.
|
|
412
|
+
They look like this:
|
|
413
|
+
|
|
414
|
+
```js
|
|
415
|
+
{
|
|
416
|
+
name: "GFP_with_locations"
|
|
417
|
+
start: 10,
|
|
418
|
+
end: 40,
|
|
419
|
+
locations: [{
|
|
420
|
+
start: 10, //this must match the .start property of the feature,
|
|
421
|
+
end: 15
|
|
422
|
+
},
|
|
423
|
+
{
|
|
424
|
+
start: 18, end: 19
|
|
425
|
+
},
|
|
426
|
+
{
|
|
427
|
+
start: 35,
|
|
428
|
+
end: 40 //this must match the .end property of the feature,
|
|
429
|
+
}
|
|
430
|
+
]
|
|
431
|
+
}
|
|
432
|
+
```
|
|
433
|
+
|
|
434
|
+
# Protein Editor
|
|
435
|
+
|
|
436
|
+
OVE can be set up to view and edit proteins (Amino Acid sequences) as first class citizens.
|
|
437
|
+
The protein editor can be seen here: http://teselagen.github.io/openVectorEditor/#/Editor?moleculeType=Protein
|
|
438
|
+
|
|
439
|
+
The editor supports Amino Acid sequences as well as DNA sequences!
|
|
440
|
+
Protein sequence mode is enabled by calling updateEditor with a protein sequenceData object:
|
|
441
|
+
|
|
442
|
+
```js
|
|
443
|
+
updateEditor(store, "DemoEditor", {
|
|
444
|
+
sequenceData: tidyUpSequenceData(exampleProteinData, {
|
|
445
|
+
convertAnnotationsFromAAIndices: true
|
|
446
|
+
})
|
|
447
|
+
});
|
|
448
|
+
```
|
|
449
|
+
|
|
450
|
+
the protein sequenceData object should look like so
|
|
451
|
+
|
|
452
|
+
```js
|
|
453
|
+
{
|
|
454
|
+
isProtein: true
|
|
455
|
+
//either or both .proteinSequence (aa string) or .sequence (dna string) must be provided if isProtein: true
|
|
456
|
+
//if only .sequence is provided, OVE will automatically compute the amino acids from the provided dna sequence
|
|
457
|
+
//if only .proteinSequence is provided, OVE will automatically compute the degenerate DNA sequence from the provided aa string
|
|
458
|
+
//if both .proteinSequence and .sequence are provided, then OVE will assume that the underlying
|
|
459
|
+
//dna sequence maps to the provided aa string as long as sequence.length === 3 * proteinSequence.length
|
|
460
|
+
proteinSequence: "mmhlrlfcillaavs...etc"
|
|
461
|
+
sequence: "gtagagagagcca...etc" //optional!
|
|
462
|
+
//if features or parts are provided to the editor, it is assumed that they will indexed to the underlying DNA sequence (0-based inclusive) , not to the AA indices .
|
|
463
|
+
//You can use the helper util from @teselagen/sequence-utils tidyUpSequenceData to convertAnnotationsFromAAIndices if your protein data has
|
|
464
|
+
//features/parts coming in as AA-indexed
|
|
465
|
+
features: [{name: "testFeature1",
|
|
466
|
+
start: 3, //start on AA 1
|
|
467
|
+
end: 5 //end on AA 1
|
|
468
|
+
}],
|
|
469
|
+
parts: [{
|
|
470
|
+
name: "myFakePart"
|
|
471
|
+
start: 0, //start on AA 0
|
|
472
|
+
end: 11 //end on AA 3
|
|
473
|
+
}]
|
|
474
|
+
}
|
|
475
|
+
```
|
|
476
|
+
|
|
477
|
+
The usual onSave, onCopy, onCut handlers will now come back with a .proteinSequence field.
|
|
478
|
+
You'll need to save/manipulate the protein sequence data however you do for dna sequences.
|
|
479
|
+
|
|
480
|
+
certain dna specific tools and annotations are automatically disabled when isProtein=true :
|
|
481
|
+
|
|
482
|
+
- primers
|
|
483
|
+
- orfs
|
|
484
|
+
- translations
|
|
485
|
+
- cutsites
|
|
486
|
+
- sequence digestions
|
|
487
|
+
- ...etc
|
|
488
|
+
|
|
489
|
+
# Alignments
|
|
490
|
+
|
|
491
|
+
## Integrating your own alignment data (only necessary if not using the built in alignment creation tool)
|
|
492
|
+
|
|
493
|
+
Add a panel to the panelsShown prop like so:
|
|
494
|
+
|
|
495
|
+
```js
|
|
496
|
+
const alignment = window.createAlignmentView(this.node, {
|
|
497
|
+
id: "jbeiAlignment1", //give your alignment a unique id
|
|
498
|
+
////optional! Use if you want a pairwise alignment:
|
|
499
|
+
pairwiseAlignments: [ // this is an array of [referenceSequence, alignedSequence]
|
|
500
|
+
[
|
|
501
|
+
{ //reference sequence must come first!
|
|
502
|
+
sequenceData: {
|
|
503
|
+
id: "FWER1231", //every sequenceData and alignmentData should have a unique id
|
|
504
|
+
name: "GFPuv58",
|
|
505
|
+
sequence: "ttgagggg",
|
|
506
|
+
features: [{id: "feat1", start: 2, end:4, name: "GFP CDS"}]
|
|
507
|
+
},
|
|
508
|
+
alignmentData: {
|
|
509
|
+
sequence: "ttgag--ggg--" //this length should be the same as the below alignmentData length!
|
|
510
|
+
}
|
|
511
|
+
},{ //aligned sequence must come second!
|
|
512
|
+
sequenceData: {
|
|
513
|
+
name: "GFPuv58",
|
|
514
|
+
sequence: "gagccgggtt"
|
|
515
|
+
},
|
|
516
|
+
alignmentData: {
|
|
517
|
+
sequence: "--gagccgggtt" //this length should be the same as the above alignmentData length!
|
|
518
|
+
}
|
|
519
|
+
}
|
|
520
|
+
]
|
|
521
|
+
[
|
|
522
|
+
{Alignment Track Data Here}, //reference sequence track (see Data Model below for specs)
|
|
523
|
+
{Alignment Track Data Here}, //aligned sequence track (see Data Model below for specs)
|
|
524
|
+
],
|
|
525
|
+
[
|
|
526
|
+
{Alignment Track Data Here}, //see Data Model below for specs
|
|
527
|
+
{Alignment Track Data Here},
|
|
528
|
+
],
|
|
529
|
+
]
|
|
530
|
+
////optional! Use if you want a multi-seq alignment:
|
|
531
|
+
alignmentTracks: [
|
|
532
|
+
{Alignment Track Data Here}, //see Data Model below for specs
|
|
533
|
+
{Alignment Track Data Here},
|
|
534
|
+
{Alignment Track Data Here},
|
|
535
|
+
],
|
|
536
|
+
//additional view options:
|
|
537
|
+
|
|
538
|
+
"alignmentAnnotationVisibility": {
|
|
539
|
+
"features": true,
|
|
540
|
+
"translations": false,
|
|
541
|
+
"parts": true,
|
|
542
|
+
"orfs": true,
|
|
543
|
+
"orfTranslations": false,
|
|
544
|
+
"axis": true,
|
|
545
|
+
"cutsites": false,
|
|
546
|
+
"primers": true,
|
|
547
|
+
"reverseSequence": false,
|
|
548
|
+
"fivePrimeThreePrimeHints": true,
|
|
549
|
+
"axisNumbers": true
|
|
550
|
+
},
|
|
551
|
+
"alignmentAnnotationLabelVisibility": {
|
|
552
|
+
"features": true,
|
|
553
|
+
"parts": true,
|
|
554
|
+
"cutsites": false
|
|
555
|
+
},
|
|
556
|
+
});
|
|
557
|
+
|
|
558
|
+
```
|
|
559
|
+
|
|
560
|
+
### Accessing the alignment state:
|
|
561
|
+
|
|
562
|
+
```js
|
|
563
|
+
const currentAlignmentState = alignment.getState();
|
|
564
|
+
//you can view various properties of the alignment such as the selection layer using alignment.getState()
|
|
565
|
+
console.info(currentAlignmentState.selectionLayer);
|
|
566
|
+
```
|
|
567
|
+
|
|
568
|
+
## Alignment Track Data Model
|
|
569
|
+
|
|
570
|
+
Note: `alignmentData.sequence` is assumed to be the same length for EVERY track within an alignment run or a pairwise alignment sub-alignment!
|
|
571
|
+
|
|
572
|
+
`alignmentData` can contain "-" characters, whereas sequenceData should not. Sequence Data is the "raw" data of the sequence being aligned with features/parts/etc.
|
|
573
|
+
|
|
574
|
+
```js
|
|
575
|
+
{
|
|
576
|
+
//JBEI sequence 'GFPuv58'
|
|
577
|
+
// chromatogramData: ab1ParsedGFPuv58,
|
|
578
|
+
sequenceData: {
|
|
579
|
+
id: "2",
|
|
580
|
+
name: "GFPuv58",
|
|
581
|
+
sequence:
|
|
582
|
+
"GTTCAATGCTTTTCCCGTTATCCGGATCATATGAAACGGCATGACTTTTTCAAGAGTGCCATGCCCGAAGGTTATGTACAGGAACGCACTATATCTTTCAAAGATGACGGGAACTACAAGACGCGTGCTGAAGTCAAGTTTGAAGGTGATACCCTTGTTAATCGTATCGAGTT"
|
|
583
|
+
},
|
|
584
|
+
alignmentData: {
|
|
585
|
+
id: "2",
|
|
586
|
+
sequence:
|
|
587
|
+
"GTTCAA--TGCTTTTCCCGTTATCCGGATCATATGAAACGGCATGACTTTTTCAAGAGTGCCATGCCCGAAGGTTATGTACA---GGAACGCACTATATCTTTCAAAGATGACGGGAACTACAAGACGCGTGCTGAAGTCAAGTTTGAAGGTGATAC--CCTTGTTAATCGTATCGAGTT--"
|
|
588
|
+
}
|
|
589
|
+
}
|
|
590
|
+
```
|
|
591
|
+
|
|
592
|
+
### Chromatogram Data
|
|
593
|
+
|
|
594
|
+
```
|
|
595
|
+
"chromatogramData": { //only if parsing in an ab1 file
|
|
596
|
+
"aTrace": [], //same as cTrace but for a
|
|
597
|
+
"tTrace": [], //same as cTrace but for t
|
|
598
|
+
"gTrace": [], //same as cTrace but for g
|
|
599
|
+
"cTrace": [0,0,0,1,3,5,11,24,56,68,54,30,21,3,1,4,1,0,0, ...etc], //heights of the curve spaced 1 per x position (aka if the cTrace.length === 1000, then the max basePos can be is 1000)
|
|
600
|
+
"basePos": [33, 46, 55,], //x position of the bases (can be unevenly spaced)
|
|
601
|
+
"baseCalls": ["A","T", ...etc],
|
|
602
|
+
"qualNums": [], //or undefined if no qualNums are detected on the file
|
|
603
|
+
},
|
|
604
|
+
```
|
|
605
|
+
|
|
606
|
+
# VersionHistoryView
|
|
607
|
+
|
|
608
|
+
## Flavors of use (aka Embedded in the Editor vs Standalone and UMD vs React):
|
|
609
|
+
|
|
610
|
+
Can be used on its own (must pass additional props):
|
|
611
|
+
|
|
612
|
+
```js
|
|
613
|
+
<VersionHistoryView
|
|
614
|
+
{...{
|
|
615
|
+
getSequenceAtVersion,
|
|
616
|
+
getVersionList,
|
|
617
|
+
onSave,
|
|
618
|
+
exitVersionHistoryView,
|
|
619
|
+
getCurrentSequenceData
|
|
620
|
+
}}
|
|
621
|
+
/>;
|
|
622
|
+
//or as a UMD module:
|
|
623
|
+
window.createVersionHistoryView({
|
|
624
|
+
getSequenceAtVersion,
|
|
625
|
+
getVersionList,
|
|
626
|
+
onSave,
|
|
627
|
+
exitVersionHistoryView,
|
|
628
|
+
getCurrentSequenceData
|
|
629
|
+
});
|
|
630
|
+
```
|
|
631
|
+
|
|
632
|
+
or as part of the Editor/createVectorEditor
|
|
633
|
+
|
|
634
|
+
```js
|
|
635
|
+
<Editor
|
|
636
|
+
{...{
|
|
637
|
+
getSequenceAtVersion,
|
|
638
|
+
getVersionList,
|
|
639
|
+
onSave,
|
|
640
|
+
ToolBarProps: { toolList: ["versionHistoryTool", ...otherTools] }
|
|
641
|
+
}}
|
|
642
|
+
/>;
|
|
643
|
+
//or as a UMD module:
|
|
644
|
+
window.createVectorEditor({
|
|
645
|
+
getSequenceAtVersion,
|
|
646
|
+
getVersionList,
|
|
647
|
+
onSave,
|
|
648
|
+
ToolBarProps: { toolList: ["versionHistoryTool", ...otherTools] }
|
|
649
|
+
});
|
|
650
|
+
```
|
|
651
|
+
|
|
652
|
+
## API:
|
|
653
|
+
|
|
654
|
+
#### getSequenceAtVersion
|
|
655
|
+
|
|
656
|
+
`(versionId) => teselagenSequenceData`
|
|
657
|
+
|
|
658
|
+
#### getVersionList
|
|
659
|
+
|
|
660
|
+
`() => [{ versionId: "51241", dateChanged: "12/30/1990", editedBy: "Hector Plahar", revisionType: "Feature Add"}]`
|
|
661
|
+
|
|
662
|
+
#### onSave [optional] (not necessary unless using the standalone VersionHistoryView)
|
|
663
|
+
|
|
664
|
+
`(event, sequenceDataToSave, editorState, onSuccessCallback) => { same onSave handler as normal } `
|
|
665
|
+
|
|
666
|
+
#### exitVersionHistoryView [optional] (not necessary unless using the standalone VersionHistoryView)
|
|
667
|
+
|
|
668
|
+
`() => {} `
|
|
669
|
+
|
|
670
|
+
#### getCurrentSequenceData [optional] (not necessary unless using the standalone VersionHistoryView)
|
|
671
|
+
|
|
672
|
+
`() => teselagenSequenceData //called upon initialization `
|
|
673
|
+
|
|
674
|
+
# Addons
|
|
675
|
+
|
|
676
|
+
## Auto annotate addon:
|
|
677
|
+
|
|
678
|
+
https://github.com/TeselaGen/openVectorEditor/tree/master/addons/AutoAnnotate
|
|
679
|
+
|
|
680
|
+
# Implementing Autosave functionality
|
|
681
|
+
|
|
682
|
+
pass shouldAutosave=true as a prop and in the onSave() handler, make sure to return a promise so that the UI responds correctly with a spinner indicating saving is in progress
|
|
683
|
+
|
|
684
|
+
# Development:
|
|
685
|
+
|
|
686
|
+
## Prerequisites
|
|
687
|
+
|
|
688
|
+
[Node.js](http://nodejs.org/) >= v8 must be installed.
|
|
689
|
+
download the latest yarn
|
|
690
|
+
|
|
691
|
+
## Outside Developer Set Up Steps
|
|
692
|
+
|
|
693
|
+
```
|
|
694
|
+
IMPORTANT! First make sure you make an issue for whatever it is you'd like to see changed!
|
|
695
|
+
fork your own branch of tg-oss from https://github.com/TeselaGen/tg-oss
|
|
696
|
+
cd tg-oss
|
|
697
|
+
yarn
|
|
698
|
+
yarn nx run ove:start #this will start the demo app
|
|
699
|
+
navigate to the output url (usually http://127.0.0.1:8805)
|
|
700
|
+
```
|
|
701
|
+
|
|
702
|
+
modify the tg-oss code base to fix/build whatever it is you're trying to do
|
|
703
|
+
set up a demo for your fix/feature if applicable in demo/src/EditorDemo/index.js
|
|
704
|
+
set up a cypress test for your fix/feature if applicable
|
|
705
|
+
you can run the cypress dev tool by running
|
|
706
|
+
|
|
707
|
+
```
|
|
708
|
+
yarn nx run ove:launch-e2e #this will run the cypress tests
|
|
709
|
+
```
|
|
710
|
+
|
|
711
|
+
you can view existing cypress tests in the packages/ove/cypress/e2e folder
|
|
712
|
+
you can either add your test to an existing cypress file or make a new test file
|
|
713
|
+
|
|
714
|
+
once you're satisfied, make a pull request back to tg-oss and mention @tnrich
|
|
715
|
+
|
|
716
|
+
```
|
|
717
|
+
|
|
718
|
+
```
|