waveorder 2.2.0rc0__tar.gz → 2.2.1__tar.gz
This diff represents the content of publicly available package versions that have been released to one of the supported registries. The information contained in this diff is provided for informational purposes only and reflects changes between package versions as they appear in their respective public registries.
- waveorder-2.2.1/.github/workflows/pr.yml +101 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/.gitignore +1 -0
- waveorder-2.2.1/.pre-commit-config.yaml +34 -0
- waveorder-2.2.1/CONTRIBUTING.md +62 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/PKG-INFO +72 -31
- waveorder-2.2.1/README.md +118 -0
- waveorder-2.2.1/examples/README.md +10 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/documentation/PTI_experiment/PTI_Experiment_Recon3D_anisotropic_target_small.py +65 -43
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/maintenance/PTI_simulation/PTI_Simulation_Forward_2D3D.py +13 -16
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/maintenance/PTI_simulation/PTI_Simulation_Recon2D.py +10 -14
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/maintenance/PTI_simulation/PTI_Simulation_Recon3D.py +14 -16
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/maintenance/QLIPP_simulation/2D_QLIPP_forward.py +6 -9
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/maintenance/QLIPP_simulation/2D_QLIPP_recon.py +9 -11
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/models/README.md +4 -2
- waveorder-2.2.1/examples/models/inplane_oriented_thick_pol3d_vector.py +92 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/models/isotropic_thin_3d.py +1 -1
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/models/phase_thick_3d.py +1 -1
- waveorder-2.2.1/examples/visuals/plot_greens_tensor.py +141 -0
- waveorder-2.2.1/examples/visuals/plot_vector_transfer_function_support.py +246 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/pyproject.toml +4 -3
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/tests/models/test_isotropic_fluorescent_thick_3d.py +0 -1
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/tests/models/test_phase_thick_3d.py +3 -3
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/tests/test_examples.py +3 -2
- waveorder-2.2.1/tests/test_filter.py +57 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/tests/test_focus_estimator.py +2 -1
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/tests/test_optics.py +3 -2
- waveorder-2.2.1/tests/test_sampling.py +10 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/tests/test_util.py +21 -2
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder/_version.py +9 -4
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder/background_estimator.py +2 -2
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder/correction.py +1 -1
- waveorder-2.2.1/waveorder/filter.py +206 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder/focus.py +5 -3
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder/models/inplane_oriented_thick_pol3d.py +16 -12
- waveorder-2.2.1/waveorder/models/inplane_oriented_thick_pol3d_vector.py +320 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder/models/isotropic_fluorescent_thick_3d.py +85 -38
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder/models/isotropic_thin_3d.py +96 -31
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder/models/phase_thick_3d.py +106 -45
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder/optics.py +233 -26
- waveorder-2.2.1/waveorder/reconstruct.py +28 -0
- waveorder-2.2.1/waveorder/sampling.py +94 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder/stokes.py +4 -3
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder/util.py +61 -9
- waveorder-2.2.0rc0/waveorder/visual.py → waveorder-2.2.1/waveorder/visuals/jupyter_visuals.py +19 -26
- waveorder-2.2.1/waveorder/visuals/matplotlib_visuals.py +335 -0
- waveorder-2.2.1/waveorder/visuals/napari_visuals.py +76 -0
- waveorder-2.2.1/waveorder/visuals/utils.py +30 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder/waveorder_reconstructor.py +18 -16
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder/waveorder_simulator.py +6 -6
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder.egg-info/PKG-INFO +72 -31
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder.egg-info/SOURCES.txt +18 -4
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder.egg-info/requires.txt +7 -2
- waveorder-2.2.0rc0/README.md +0 -82
- waveorder-2.2.0rc0/examples/README.md +0 -7
- waveorder-2.2.0rc0/examples/models/isotropic_thin_3d_resolution.py +0 -88
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/.git-blame-ignore-revs +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/.github/workflows/pytests.yml +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/CITATION.cff +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/LICENSE +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/docs/valuable-prs/2023-02-27.110.pr.open.md +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/documentation/PTI_experiment/PTI_Experiment_Recon3D_anisotropic_target_small.pdf +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/documentation/PTI_experiment/PTI_full_FOV_anisotropic_target.ipynb +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/documentation/PTI_experiment/PTI_full_FOV_cardiac_muscle.ipynb +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/documentation/PTI_experiment/PTI_full_FOV_cardiomyocyte_infected_1.ipynb +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/documentation/PTI_experiment/PTI_full_FOV_cardiomyocyte_infected_2.ipynb +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/documentation/PTI_experiment/PTI_full_FOV_cardiomyocyte_mock.ipynb +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/documentation/PTI_experiment/PTI_full_FOV_human_uterus.ipynb +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/documentation/PTI_experiment/PTI_full_FOV_mouse_brain_aco.ipynb +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/documentation/PTI_experiment/README.md +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/documentation/QLIPP_experiment/2D_QLIPP_recon_experiment.ipynb +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/documentation/QLIPP_experiment/3D_QLIPP_recon_experiment.ipynb +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/documentation/README.md +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/documentation/fluorescence_deconvolution/fluorescence_deconv.ipynb +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/maintenance/PTI_simulation/PTI_formulation.html +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/maintenance/PTI_simulation/README.md +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/maintenance/README.md +0 -0
- /waveorder-2.2.0rc0/examples/models/inplane_oriented_thick_pol3D.py → /waveorder-2.2.1/examples/models/inplane_oriented_thick_pol3d.py +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/examples/models/isotropic_fluorescent_thick_3d.py +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/readme.png +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/setup.cfg +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/tests/__init__.py +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/tests/conftest.py +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/tests/models/test_inplane_oriented_thick_pol3D.py +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/tests/models/test_isotropic_thin_3d.py +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/tests/test_correction.py +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/tests/test_stokes.py +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder/__init__.py +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder.egg-info/dependency_links.txt +0 -0
- {waveorder-2.2.0rc0 → waveorder-2.2.1}/waveorder.egg-info/top_level.txt +0 -0
|
@@ -0,0 +1,101 @@
|
|
|
1
|
+
# This workflow will install Python dependencies, run tests and lint with a single version of Python
|
|
2
|
+
# For more information see: https://help.github.com/actions/language-and-framework-guides/using-python-with-github-actions
|
|
3
|
+
|
|
4
|
+
name: lint, style, and tests
|
|
5
|
+
|
|
6
|
+
on:
|
|
7
|
+
pull_request:
|
|
8
|
+
branches:
|
|
9
|
+
- main
|
|
10
|
+
|
|
11
|
+
jobs:
|
|
12
|
+
style:
|
|
13
|
+
name: Style Check
|
|
14
|
+
runs-on: ubuntu-latest
|
|
15
|
+
|
|
16
|
+
strategy:
|
|
17
|
+
matrix:
|
|
18
|
+
python-version: ["3.10"]
|
|
19
|
+
|
|
20
|
+
steps:
|
|
21
|
+
- uses: actions/checkout@v3
|
|
22
|
+
- name: Set up Python
|
|
23
|
+
uses: actions/setup-python@v4
|
|
24
|
+
with:
|
|
25
|
+
python-version: ${{ matrix.python-version }}
|
|
26
|
+
- name: Install dependencies
|
|
27
|
+
run: |
|
|
28
|
+
python -m pip install --upgrade pip
|
|
29
|
+
pip install black==25.1.0
|
|
30
|
+
- name: Check code styling with Black
|
|
31
|
+
run: |
|
|
32
|
+
black --diff -S -t py310 waveorder
|
|
33
|
+
black --check -S -t py310 waveorder
|
|
34
|
+
|
|
35
|
+
# lint:
|
|
36
|
+
# name: Lint Check
|
|
37
|
+
# runs-on: ubuntu-latest
|
|
38
|
+
|
|
39
|
+
# strategy:
|
|
40
|
+
# matrix:
|
|
41
|
+
# python-version: ["3.10"]
|
|
42
|
+
|
|
43
|
+
# steps:
|
|
44
|
+
# - uses: actions/checkout@v3
|
|
45
|
+
# - name: Set up Python
|
|
46
|
+
# uses: actions/setup-python@v4
|
|
47
|
+
# with:
|
|
48
|
+
# python-version: ${{ matrix.python-version }}
|
|
49
|
+
# - name: Install dependencies
|
|
50
|
+
# run: |
|
|
51
|
+
# python -m pip install --upgrade pip
|
|
52
|
+
# pip install flake8
|
|
53
|
+
# - name: Check code with Flake8
|
|
54
|
+
# # E203 conflicts with black
|
|
55
|
+
# run: |
|
|
56
|
+
# flake8 waveorder --extend-ignore=E203
|
|
57
|
+
|
|
58
|
+
isort:
|
|
59
|
+
name: isort Check
|
|
60
|
+
runs-on: ubuntu-latest
|
|
61
|
+
|
|
62
|
+
strategy:
|
|
63
|
+
matrix:
|
|
64
|
+
python-version: ["3.10"]
|
|
65
|
+
|
|
66
|
+
steps:
|
|
67
|
+
- uses: actions/checkout@v3
|
|
68
|
+
- name: Set up Python
|
|
69
|
+
uses: actions/setup-python@v4
|
|
70
|
+
with:
|
|
71
|
+
python-version: ${{ matrix.python-version }}
|
|
72
|
+
- name: Install dependencies
|
|
73
|
+
run: |
|
|
74
|
+
python -m pip install --upgrade pip
|
|
75
|
+
pip install isort
|
|
76
|
+
- name: Check code with isort
|
|
77
|
+
run: |
|
|
78
|
+
isort --check waveorder
|
|
79
|
+
|
|
80
|
+
tests:
|
|
81
|
+
needs: [style, isort] # lint
|
|
82
|
+
runs-on: ubuntu-latest
|
|
83
|
+
strategy:
|
|
84
|
+
matrix:
|
|
85
|
+
python-version: ["3.10", "3.11", "3.12"]
|
|
86
|
+
|
|
87
|
+
steps:
|
|
88
|
+
- uses: actions/checkout@v3
|
|
89
|
+
|
|
90
|
+
- uses: actions/setup-python@v4
|
|
91
|
+
with:
|
|
92
|
+
python-version: ${{ matrix.python-version }}
|
|
93
|
+
|
|
94
|
+
- name: Install dependencies
|
|
95
|
+
run: |
|
|
96
|
+
python -m pip install --upgrade pip
|
|
97
|
+
pip install ".[dev]"
|
|
98
|
+
|
|
99
|
+
- name: Test with pytest
|
|
100
|
+
run: |
|
|
101
|
+
pytest -v --cov=./ --cov-report=xml
|
|
@@ -0,0 +1,34 @@
|
|
|
1
|
+
|
|
2
|
+
repos:
|
|
3
|
+
# basic pre-commit
|
|
4
|
+
- repo: https://github.com/pre-commit/pre-commit-hooks
|
|
5
|
+
rev: v4.4.0
|
|
6
|
+
hooks:
|
|
7
|
+
- id: trailing-whitespace
|
|
8
|
+
- id: end-of-file-fixer
|
|
9
|
+
- id: check-added-large-files
|
|
10
|
+
- id: check-yaml
|
|
11
|
+
- id: check-toml
|
|
12
|
+
- id: detect-private-key
|
|
13
|
+
# sorting imports
|
|
14
|
+
- repo: https://github.com/pycqa/isort
|
|
15
|
+
rev: 5.12.0
|
|
16
|
+
hooks:
|
|
17
|
+
- id: isort
|
|
18
|
+
# syntax linting and formatting
|
|
19
|
+
- repo: https://github.com/myint/autoflake
|
|
20
|
+
rev: v2.1.1
|
|
21
|
+
hooks:
|
|
22
|
+
- id: autoflake
|
|
23
|
+
args: [--in-place, --remove-all-unused-imports,
|
|
24
|
+
--ignore-init-module-imports]
|
|
25
|
+
# - repo: https://github.com/PyCQA/flake8
|
|
26
|
+
# rev: 6.0.0
|
|
27
|
+
# hooks:
|
|
28
|
+
# - id: flake8
|
|
29
|
+
# args: [--ignore, "E203,W503", --min-python-version, '3.10']
|
|
30
|
+
# additional_dependencies: [flake8-typing-imports==1.12.0]
|
|
31
|
+
- repo: https://github.com/psf/black
|
|
32
|
+
rev: 25.1.0
|
|
33
|
+
hooks:
|
|
34
|
+
- id: black
|
|
@@ -0,0 +1,62 @@
|
|
|
1
|
+
# Contributing guide
|
|
2
|
+
|
|
3
|
+
Thanks for your interest in contributing to `waveorder`!
|
|
4
|
+
|
|
5
|
+
Please see the following steps for our workflow.
|
|
6
|
+
|
|
7
|
+
## Getting started
|
|
8
|
+
|
|
9
|
+
Please read the [README](./README.md) for an overview of the project,
|
|
10
|
+
and how you can install and use the package.
|
|
11
|
+
|
|
12
|
+
## Issues
|
|
13
|
+
|
|
14
|
+
We use [issues](https://github.com/mehta-lab/waveorder/issues) to track
|
|
15
|
+
bug reports, feature requests, and provide user support.
|
|
16
|
+
|
|
17
|
+
Before opening a new issue, please first search existing issues (including closed ones),
|
|
18
|
+
to see if there is an existing discussion about it.
|
|
19
|
+
|
|
20
|
+
### Setting up development environment
|
|
21
|
+
|
|
22
|
+
For local development, first install [Git](https://git-scm.com/)
|
|
23
|
+
and Python with an environment management tool
|
|
24
|
+
(e.g. [miniforge](https://github.com/conda-forge/miniforge), a minimal community distribution of Conda).
|
|
25
|
+
|
|
26
|
+
If you use Conda, set up an environment with:
|
|
27
|
+
|
|
28
|
+
```sh
|
|
29
|
+
conda create -n waveorder-dev python=3.10
|
|
30
|
+
conda activate waveorder-dev
|
|
31
|
+
```
|
|
32
|
+
|
|
33
|
+
If you have push permission to the repository,
|
|
34
|
+
clone the repository (the code blocks below are shell commands):
|
|
35
|
+
|
|
36
|
+
```sh
|
|
37
|
+
cd # to the directory you want to work in
|
|
38
|
+
git clone https://github.com/mehta-lab/waveorder.git
|
|
39
|
+
```
|
|
40
|
+
|
|
41
|
+
Otherwise, you can follow [these instructions](https://docs.github.com/en/get-started/quickstart/fork-a-repo)
|
|
42
|
+
to [fork](https://github.com/mehta-lab/waveorder/fork) the repository.
|
|
43
|
+
|
|
44
|
+
Then install the package in editable mode with the development dependencies:
|
|
45
|
+
|
|
46
|
+
```sh
|
|
47
|
+
cd waveorder/ # or the renamed project root directory
|
|
48
|
+
pip install -e ".[dev]"
|
|
49
|
+
```
|
|
50
|
+
|
|
51
|
+
Then make the changes and [track them with Git](https://docs.github.com/en/get-started/using-git/about-git#example-contribute-to-an-existing-repository).
|
|
52
|
+
|
|
53
|
+
|
|
54
|
+
### Code style
|
|
55
|
+
|
|
56
|
+
We use [pre-commit](https://pre-commit.com/) to sort imports with [isort](https://github.com/PyCQA/isort) and format code with [black](https://black.readthedocs.io/en/stable/) automatically prior to each commit. To minimize test errors when submitting pull requests, please install pre-commit in your environment as follows:
|
|
57
|
+
|
|
58
|
+
```bash
|
|
59
|
+
pre-commit install
|
|
60
|
+
```
|
|
61
|
+
|
|
62
|
+
When these packages are executed within the project root directory, they should automatically use the [project settings](./pyproject.toml).
|
|
@@ -1,6 +1,6 @@
|
|
|
1
|
-
Metadata-Version: 2.
|
|
1
|
+
Metadata-Version: 2.4
|
|
2
2
|
Name: waveorder
|
|
3
|
-
Version: 2.2.
|
|
3
|
+
Version: 2.2.1
|
|
4
4
|
Summary: Wave-optical simulations and deconvolution of optical properties
|
|
5
5
|
Author-email: CZ Biohub SF <compmicro@czbiohub.org>
|
|
6
6
|
Maintainer-email: Talon Chandler <talon.chandler@czbiohub.org>, Shalin Mehta <shalin.mehta@czbiohub.org>
|
|
@@ -53,59 +53,101 @@ Classifier: Operating System :: MacOS
|
|
|
53
53
|
Requires-Python: >=3.10
|
|
54
54
|
Description-Content-Type: text/markdown
|
|
55
55
|
License-File: LICENSE
|
|
56
|
-
Requires-Dist: numpy
|
|
56
|
+
Requires-Dist: numpy>=1.24
|
|
57
57
|
Requires-Dist: matplotlib>=3.1.1
|
|
58
58
|
Requires-Dist: scipy>=1.3.0
|
|
59
59
|
Requires-Dist: pywavelets>=1.1.1
|
|
60
60
|
Requires-Dist: ipywidgets>=7.5.1
|
|
61
|
-
Requires-Dist: torch>=2.
|
|
61
|
+
Requires-Dist: torch>=2.4.1
|
|
62
62
|
Provides-Extra: dev
|
|
63
63
|
Requires-Dist: pytest; extra == "dev"
|
|
64
64
|
Requires-Dist: pytest-cov; extra == "dev"
|
|
65
|
+
Requires-Dist: black==25.1.0; extra == "dev"
|
|
66
|
+
Provides-Extra: examples
|
|
67
|
+
Requires-Dist: napari[all]; extra == "examples"
|
|
68
|
+
Requires-Dist: jupyter; extra == "examples"
|
|
69
|
+
Dynamic: license-file
|
|
65
70
|
|
|
66
71
|
# waveorder
|
|
67
72
|
|
|
68
|
-

|
|
69
|
-
[](https://pepy.tech/project/waveorder)
|
|
70
73
|
[](https://pypi.org/project/waveorder)
|
|
71
|
-
[](https://pypistats.org/packages/waveorder)
|
|
75
|
+
[](https://pepy.tech/project/waveorder)
|
|
76
|
+
[](https://github.com/mehta-lab/waveorder/graphs/contributors)
|
|
77
|
+

|
|
78
|
+

|
|
79
|
+

|
|
80
|
+
|
|
72
81
|
|
|
73
82
|
This computational imaging library enables wave-optical simulation and reconstruction of optical properties that report microscopic architectural order.
|
|
74
83
|
|
|
75
|
-
## Computational label-
|
|
84
|
+
## Computational label-agnostic imaging
|
|
76
85
|
|
|
77
|
-
|
|
86
|
+
https://github.com/user-attachments/assets/4f9969e5-94ce-4e08-9f30-68314a905db6
|
|
78
87
|
|
|
79
|
-
|
|
88
|
+
`waveorder` enables simulations and reconstructions of label-agnostic microscopy data as described in the following [preprint](https://arxiv.org/abs/2412.09775)
|
|
89
|
+
<details>
|
|
90
|
+
<summary> Chandler et al. 2024 </summary>
|
|
91
|
+
<pre><code>
|
|
92
|
+
@article{chandler_2024,
|
|
93
|
+
author = {Chandler, Talon and Hirata-Miyasaki, Eduardo and Ivanov, Ivan E. and Liu, Ziwen and Sundarraman, Deepika and Ryan, Allyson Quinn and Jacobo, Adrian and Balla, Keir and Mehta, Shalin B.},
|
|
94
|
+
title = {waveOrder: generalist framework for label-agnostic computational microscopy},
|
|
95
|
+
journal = {arXiv},
|
|
96
|
+
year = {2024},
|
|
97
|
+
month = dec,
|
|
98
|
+
eprint = {2412.09775},
|
|
99
|
+
doi = {10.48550/arXiv.2412.09775}
|
|
100
|
+
}
|
|
101
|
+
</code></pre>
|
|
102
|
+
</details>
|
|
80
103
|
|
|
81
|
-
|
|
82
|
-
L.-H. Yeh, I. E. Ivanov, B. B. Chhun, S.-M. Guo, E. Hashemi, J. R. Byrum, J. A. Pérez-Bermejo, H. Wang, Y. Yu, P. G. Kazansky, B. R. Conklin, M. H. Han, and S. B. Mehta, "uPTI: uniaxial permittivity tensor imaging of intrinsic density and anisotropy," bioRxiv 2020.12.15.422951 (2020).
|
|
83
|
-
```
|
|
104
|
+
Specifically, `waveorder` enables simulation and reconstruction of 2D or 3D:
|
|
84
105
|
|
|
85
|
-
|
|
106
|
+
1. __phase, projected retardance, and in-plane orientation__ from a polarization-diverse volumetric brightfield acquisition ([QLIPP](https://elifesciences.org/articles/55502)),
|
|
86
107
|
|
|
87
|
-
|
|
108
|
+
2. __phase__ from a volumetric brightfield acquisition ([2D phase](https://www.osapublishing.org/ao/abstract.cfm?uri=ao-54-28-8566)/[3D phase](https://www.osapublishing.org/ao/abstract.cfm?uri=ao-57-1-a205)),
|
|
88
109
|
|
|
89
|
-
|
|
110
|
+
3. __phase__ from an illumination-diverse volumetric acquisition ([2D](https://www.osapublishing.org/oe/fulltext.cfm?uri=oe-23-9-11394&id=315599)/[3D](https://www.osapublishing.org/boe/fulltext.cfm?uri=boe-7-10-3940&id=349951) differential phase contrast),
|
|
90
111
|
|
|
91
|
-
|
|
112
|
+
4. __fluorescence density__ from a widefield volumetric fluorescence acquisition (fluorescence deconvolution).
|
|
92
113
|
|
|
93
|
-
|
|
114
|
+
The [examples](https://github.com/mehta-lab/waveorder/tree/main/examples) demonstrate simulations and reconstruction for 2D QLIPP, 3D PODT, 3D fluorescence deconvolution, and 2D/3D PTI methods.
|
|
94
115
|
|
|
95
|
-
|
|
116
|
+
If you are interested in deploying QLIPP, phase from brightfield, or fluorescence deconvolution for label-agnostic imaging at scale, checkout our [napari plugin](https://www.napari-hub.org/plugins/recOrder-napari), [`recOrder-napari`](https://github.com/mehta-lab/recOrder).
|
|
96
117
|
|
|
118
|
+
## Permittivity tensor imaging
|
|
97
119
|
|
|
98
|
-
|
|
120
|
+
Additionally, `waveorder` enabled the development of a new label-free imaging method, __permittivity tensor imaging (PTI)__, that measures density and 3D orientation of biomolecules with diffraction-limited resolution. These measurements are reconstructed from polarization-resolved images acquired with a sequence of oblique illuminations.
|
|
99
121
|
|
|
100
|
-
|
|
122
|
+
The acquisition, calibration, background correction, reconstruction, and applications of PTI are described in the following [paper](https://doi.org/10.1101/2020.12.15.422951) published in Nature Methods:
|
|
101
123
|
|
|
102
|
-
|
|
124
|
+
<details>
|
|
125
|
+
<summary> Yeh et al. 2024 </summary>
|
|
126
|
+
<pre><code>
|
|
127
|
+
@article{yeh_2024,
|
|
128
|
+
author = {Yeh, Li-Hao and Ivanov, Ivan E. and Chandler, Talon and Byrum, Janie R. and Chhun, Bryant B. and Guo, Syuan-Ming and Foltz, Cameron and Hashemi, Ezzat and Perez-Bermejo, Juan A. and Wang, Huijun and Yu, Yanhao and Kazansky, Peter G. and Conklin, Bruce R. and Han, May H. and Mehta, Shalin B.},
|
|
129
|
+
title = {Permittivity tensor imaging: modular label-free imaging of 3D dry mass and 3D orientation at high resolution},
|
|
130
|
+
journal = {Nature Methods},
|
|
131
|
+
volume = {21},
|
|
132
|
+
number = {7},
|
|
133
|
+
pages = {1257--1274},
|
|
134
|
+
year = {2024},
|
|
135
|
+
month = jul,
|
|
136
|
+
issn = {1548-7105},
|
|
137
|
+
publisher = {Nature Publishing Group},
|
|
138
|
+
doi = {10.1038/s41592-024-02291-w}
|
|
139
|
+
}
|
|
140
|
+
</code></pre>
|
|
141
|
+
</details>
|
|
103
142
|
|
|
104
|
-
|
|
143
|
+
PTI provides volumetric reconstructions of mean permittivity ($\propto$ material density), differential permittivity ($\propto$ material anisotropy), 3D orientation, and optic sign. The following figure summarizes PTI acquisition and reconstruction with a small optical section of the mouse brain tissue:
|
|
144
|
+
|
|
145
|
+
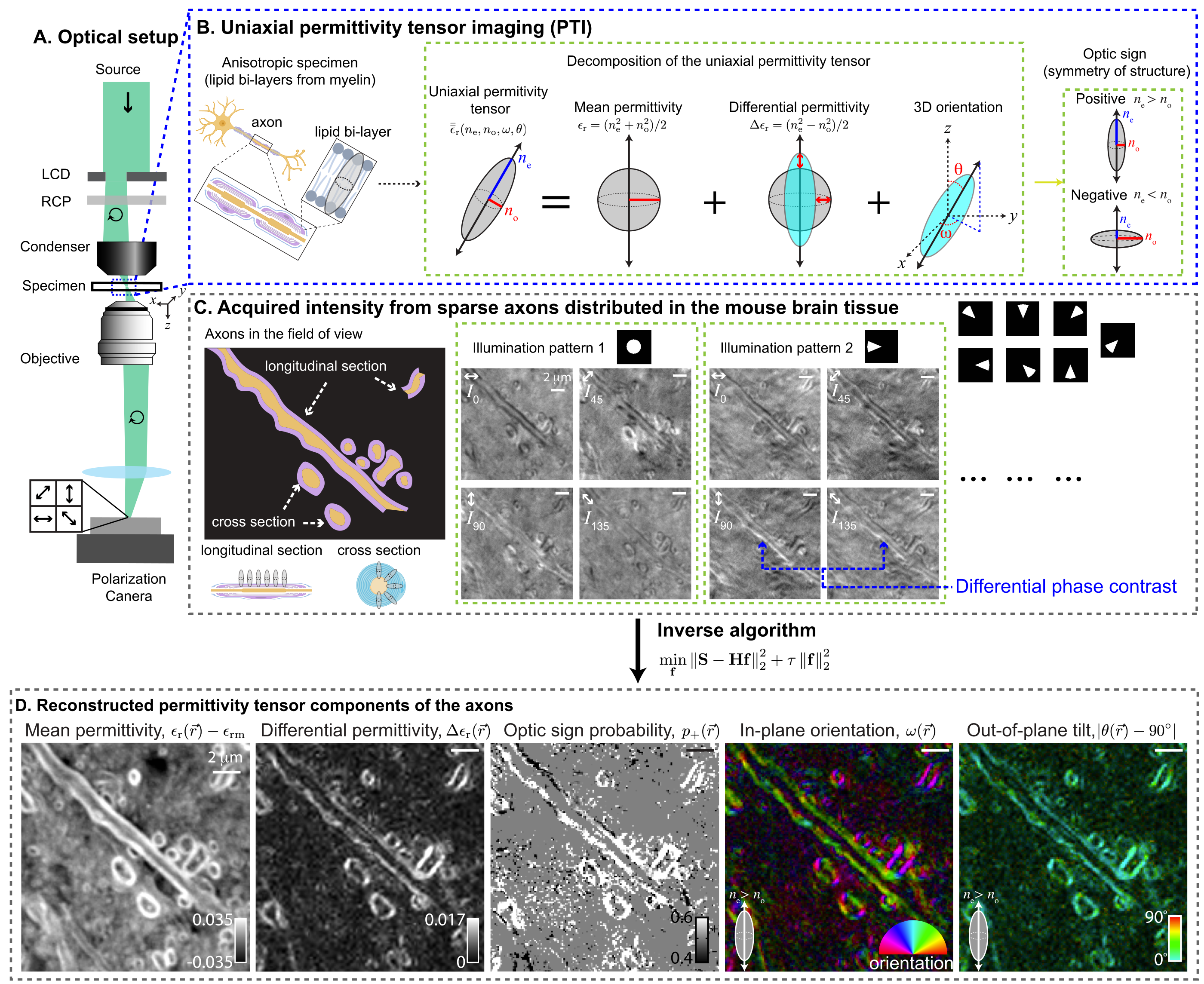
|
|
105
146
|
|
|
106
|
-
|
|
147
|
+
## Examples
|
|
148
|
+
The [examples](https://github.com/mehta-lab/waveorder/tree/main/examples) illustrate simulations and reconstruction for 2D QLIPP, 3D phase from brightfield, and 2D/3D PTI methods.
|
|
107
149
|
|
|
108
|
-
|
|
150
|
+
If you are interested in deploying QLIPP or phase from brightbrield, or fluorescence deconvolution for label-agnostic imaging at scale, checkout our [napari plugin](https://www.napari-hub.org/plugins/recOrder-napari), [`recOrder-napari`](https://github.com/mehta-lab/recOrder).
|
|
109
151
|
|
|
110
152
|
## Citation
|
|
111
153
|
|
|
@@ -113,10 +155,10 @@ Please cite this repository, along with the relevant preprint or paper, if you u
|
|
|
113
155
|
|
|
114
156
|
## Installation
|
|
115
157
|
|
|
116
|
-
|
|
158
|
+
Create a virtual environment:
|
|
117
159
|
|
|
118
160
|
```sh
|
|
119
|
-
conda create -y -n waveorder python=3.
|
|
161
|
+
conda create -y -n waveorder python=3.10
|
|
120
162
|
conda activate waveorder
|
|
121
163
|
```
|
|
122
164
|
|
|
@@ -133,12 +175,11 @@ python
|
|
|
133
175
|
>>> import waveorder
|
|
134
176
|
```
|
|
135
177
|
|
|
136
|
-
(Optional)
|
|
137
|
-
|
|
178
|
+
(Optional) Install example dependencies, clone the repository, and run an example script:
|
|
138
179
|
```sh
|
|
180
|
+
pip install waveorder[examples]
|
|
139
181
|
git clone https://github.com/mehta-lab/waveorder.git
|
|
140
|
-
|
|
141
|
-
jupyter notebook ./waveorder/examples/
|
|
182
|
+
python waveorder/examples/models/phase_thick_3d.py
|
|
142
183
|
```
|
|
143
184
|
|
|
144
185
|
(M1 users) `pytorch` has [incomplete GPU support](https://github.com/pytorch/pytorch/issues/77764),
|
|
@@ -0,0 +1,118 @@
|
|
|
1
|
+
# waveorder
|
|
2
|
+
|
|
3
|
+
[](https://pypi.org/project/waveorder)
|
|
4
|
+
[](https://pypistats.org/packages/waveorder)
|
|
5
|
+
[](https://pepy.tech/project/waveorder)
|
|
6
|
+
[](https://github.com/mehta-lab/waveorder/graphs/contributors)
|
|
7
|
+

|
|
8
|
+

|
|
9
|
+

|
|
10
|
+
|
|
11
|
+
|
|
12
|
+
This computational imaging library enables wave-optical simulation and reconstruction of optical properties that report microscopic architectural order.
|
|
13
|
+
|
|
14
|
+
## Computational label-agnostic imaging
|
|
15
|
+
|
|
16
|
+
https://github.com/user-attachments/assets/4f9969e5-94ce-4e08-9f30-68314a905db6
|
|
17
|
+
|
|
18
|
+
`waveorder` enables simulations and reconstructions of label-agnostic microscopy data as described in the following [preprint](https://arxiv.org/abs/2412.09775)
|
|
19
|
+
<details>
|
|
20
|
+
<summary> Chandler et al. 2024 </summary>
|
|
21
|
+
<pre><code>
|
|
22
|
+
@article{chandler_2024,
|
|
23
|
+
author = {Chandler, Talon and Hirata-Miyasaki, Eduardo and Ivanov, Ivan E. and Liu, Ziwen and Sundarraman, Deepika and Ryan, Allyson Quinn and Jacobo, Adrian and Balla, Keir and Mehta, Shalin B.},
|
|
24
|
+
title = {waveOrder: generalist framework for label-agnostic computational microscopy},
|
|
25
|
+
journal = {arXiv},
|
|
26
|
+
year = {2024},
|
|
27
|
+
month = dec,
|
|
28
|
+
eprint = {2412.09775},
|
|
29
|
+
doi = {10.48550/arXiv.2412.09775}
|
|
30
|
+
}
|
|
31
|
+
</code></pre>
|
|
32
|
+
</details>
|
|
33
|
+
|
|
34
|
+
Specifically, `waveorder` enables simulation and reconstruction of 2D or 3D:
|
|
35
|
+
|
|
36
|
+
1. __phase, projected retardance, and in-plane orientation__ from a polarization-diverse volumetric brightfield acquisition ([QLIPP](https://elifesciences.org/articles/55502)),
|
|
37
|
+
|
|
38
|
+
2. __phase__ from a volumetric brightfield acquisition ([2D phase](https://www.osapublishing.org/ao/abstract.cfm?uri=ao-54-28-8566)/[3D phase](https://www.osapublishing.org/ao/abstract.cfm?uri=ao-57-1-a205)),
|
|
39
|
+
|
|
40
|
+
3. __phase__ from an illumination-diverse volumetric acquisition ([2D](https://www.osapublishing.org/oe/fulltext.cfm?uri=oe-23-9-11394&id=315599)/[3D](https://www.osapublishing.org/boe/fulltext.cfm?uri=boe-7-10-3940&id=349951) differential phase contrast),
|
|
41
|
+
|
|
42
|
+
4. __fluorescence density__ from a widefield volumetric fluorescence acquisition (fluorescence deconvolution).
|
|
43
|
+
|
|
44
|
+
The [examples](https://github.com/mehta-lab/waveorder/tree/main/examples) demonstrate simulations and reconstruction for 2D QLIPP, 3D PODT, 3D fluorescence deconvolution, and 2D/3D PTI methods.
|
|
45
|
+
|
|
46
|
+
If you are interested in deploying QLIPP, phase from brightfield, or fluorescence deconvolution for label-agnostic imaging at scale, checkout our [napari plugin](https://www.napari-hub.org/plugins/recOrder-napari), [`recOrder-napari`](https://github.com/mehta-lab/recOrder).
|
|
47
|
+
|
|
48
|
+
## Permittivity tensor imaging
|
|
49
|
+
|
|
50
|
+
Additionally, `waveorder` enabled the development of a new label-free imaging method, __permittivity tensor imaging (PTI)__, that measures density and 3D orientation of biomolecules with diffraction-limited resolution. These measurements are reconstructed from polarization-resolved images acquired with a sequence of oblique illuminations.
|
|
51
|
+
|
|
52
|
+
The acquisition, calibration, background correction, reconstruction, and applications of PTI are described in the following [paper](https://doi.org/10.1101/2020.12.15.422951) published in Nature Methods:
|
|
53
|
+
|
|
54
|
+
<details>
|
|
55
|
+
<summary> Yeh et al. 2024 </summary>
|
|
56
|
+
<pre><code>
|
|
57
|
+
@article{yeh_2024,
|
|
58
|
+
author = {Yeh, Li-Hao and Ivanov, Ivan E. and Chandler, Talon and Byrum, Janie R. and Chhun, Bryant B. and Guo, Syuan-Ming and Foltz, Cameron and Hashemi, Ezzat and Perez-Bermejo, Juan A. and Wang, Huijun and Yu, Yanhao and Kazansky, Peter G. and Conklin, Bruce R. and Han, May H. and Mehta, Shalin B.},
|
|
59
|
+
title = {Permittivity tensor imaging: modular label-free imaging of 3D dry mass and 3D orientation at high resolution},
|
|
60
|
+
journal = {Nature Methods},
|
|
61
|
+
volume = {21},
|
|
62
|
+
number = {7},
|
|
63
|
+
pages = {1257--1274},
|
|
64
|
+
year = {2024},
|
|
65
|
+
month = jul,
|
|
66
|
+
issn = {1548-7105},
|
|
67
|
+
publisher = {Nature Publishing Group},
|
|
68
|
+
doi = {10.1038/s41592-024-02291-w}
|
|
69
|
+
}
|
|
70
|
+
</code></pre>
|
|
71
|
+
</details>
|
|
72
|
+
|
|
73
|
+
PTI provides volumetric reconstructions of mean permittivity ($\propto$ material density), differential permittivity ($\propto$ material anisotropy), 3D orientation, and optic sign. The following figure summarizes PTI acquisition and reconstruction with a small optical section of the mouse brain tissue:
|
|
74
|
+
|
|
75
|
+

|
|
76
|
+
|
|
77
|
+
## Examples
|
|
78
|
+
The [examples](https://github.com/mehta-lab/waveorder/tree/main/examples) illustrate simulations and reconstruction for 2D QLIPP, 3D phase from brightfield, and 2D/3D PTI methods.
|
|
79
|
+
|
|
80
|
+
If you are interested in deploying QLIPP or phase from brightbrield, or fluorescence deconvolution for label-agnostic imaging at scale, checkout our [napari plugin](https://www.napari-hub.org/plugins/recOrder-napari), [`recOrder-napari`](https://github.com/mehta-lab/recOrder).
|
|
81
|
+
|
|
82
|
+
## Citation
|
|
83
|
+
|
|
84
|
+
Please cite this repository, along with the relevant preprint or paper, if you use or adapt this code. The citation information can be found by clicking "Cite this repository" button in the About section in the right sidebar.
|
|
85
|
+
|
|
86
|
+
## Installation
|
|
87
|
+
|
|
88
|
+
Create a virtual environment:
|
|
89
|
+
|
|
90
|
+
```sh
|
|
91
|
+
conda create -y -n waveorder python=3.10
|
|
92
|
+
conda activate waveorder
|
|
93
|
+
```
|
|
94
|
+
|
|
95
|
+
Install `waveorder` from PyPI:
|
|
96
|
+
|
|
97
|
+
```sh
|
|
98
|
+
pip install waveorder
|
|
99
|
+
```
|
|
100
|
+
|
|
101
|
+
Use `waveorder` in your scripts:
|
|
102
|
+
|
|
103
|
+
```sh
|
|
104
|
+
python
|
|
105
|
+
>>> import waveorder
|
|
106
|
+
```
|
|
107
|
+
|
|
108
|
+
(Optional) Install example dependencies, clone the repository, and run an example script:
|
|
109
|
+
```sh
|
|
110
|
+
pip install waveorder[examples]
|
|
111
|
+
git clone https://github.com/mehta-lab/waveorder.git
|
|
112
|
+
python waveorder/examples/models/phase_thick_3d.py
|
|
113
|
+
```
|
|
114
|
+
|
|
115
|
+
(M1 users) `pytorch` has [incomplete GPU support](https://github.com/pytorch/pytorch/issues/77764),
|
|
116
|
+
so please use `export PYTORCH_ENABLE_MPS_FALLBACK=1`
|
|
117
|
+
to allow some operators to fallback to CPU
|
|
118
|
+
if you plan to use GPU acceleration for polarization reconstruction.
|
|
@@ -0,0 +1,10 @@
|
|
|
1
|
+
`waveorder` is undergoing a significant refactor, and this `examples/` folder serves as a good place to understand the current state of the repository.
|
|
2
|
+
|
|
3
|
+
Some examples require `pip install waveorder[examples]` for `napari` and `jupyter`. Visit the [napari installation guide](https://napari.org/dev/tutorials/fundamentals/installation.html) if napari installation fails.
|
|
4
|
+
|
|
5
|
+
| Folder | Requires | Description |
|
|
6
|
+
|------------------|----------------------------|-------------------------------------------------------------------------------------------------------|
|
|
7
|
+
| `models/` | `pip install waveorder[examples]` | Demonstrates the latest functionality of `waveorder` through simulations and reconstructions using various models. |
|
|
8
|
+
| `maintenance/` | `pip install waveorder` | Examples of computational imaging methods enabled by functionality of waveorder; scripts are maintained with automated tests. |
|
|
9
|
+
| `visuals/` | `pip install waveorder[examples]` | Visualizations of transfer functions and Green's tensors. |
|
|
10
|
+
| `documentation/` | `pip install waveorder`, complete datasets | Provides examples of real-data reconstructions; serves as documentation and is not actively maintained. |
|