waveorder 2.2.0__tar.gz → 2.2.0rc0__tar.gz
This diff represents the content of publicly available package versions that have been released to one of the supported registries. The information contained in this diff is provided for informational purposes only and reflects changes between package versions as they appear in their respective public registries.
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/.gitignore +0 -1
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/PKG-INFO +30 -69
- waveorder-2.2.0rc0/README.md +82 -0
- waveorder-2.2.0rc0/examples/README.md +7 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/documentation/PTI_experiment/PTI_Experiment_Recon3D_anisotropic_target_small.py +13 -14
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/maintenance/PTI_simulation/PTI_Simulation_Forward_2D3D.py +10 -10
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/maintenance/PTI_simulation/PTI_Simulation_Recon2D.py +8 -8
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/maintenance/PTI_simulation/PTI_Simulation_Recon3D.py +11 -11
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/maintenance/QLIPP_simulation/2D_QLIPP_forward.py +3 -3
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/maintenance/QLIPP_simulation/2D_QLIPP_recon.py +7 -7
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/models/README.md +2 -4
- waveorder-2.2.0rc0/examples/models/isotropic_thin_3d_resolution.py +88 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/pyproject.toml +2 -3
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/tests/test_optics.py +1 -1
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/tests/test_util.py +0 -18
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder/_version.py +1 -1
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder/models/inplane_oriented_thick_pol3d.py +12 -12
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder/models/isotropic_fluorescent_thick_3d.py +34 -70
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder/models/isotropic_thin_3d.py +32 -94
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder/models/phase_thick_3d.py +43 -94
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder/optics.py +22 -232
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder/util.py +2 -54
- waveorder-2.2.0/waveorder/visuals/jupyter_visuals.py → waveorder-2.2.0rc0/waveorder/visual.py +6 -2
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder/waveorder_reconstructor.py +7 -8
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder.egg-info/PKG-INFO +30 -69
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder.egg-info/SOURCES.txt +4 -12
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder.egg-info/requires.txt +2 -6
- waveorder-2.2.0/README.md +0 -118
- waveorder-2.2.0/examples/README.md +0 -10
- waveorder-2.2.0/examples/models/inplane_oriented_thick_pol3d_vector.py +0 -92
- waveorder-2.2.0/examples/visuals/plot_greens_tensor.py +0 -139
- waveorder-2.2.0/examples/visuals/plot_vector_transfer_function_support.py +0 -241
- waveorder-2.2.0/tests/test_sampling.py +0 -13
- waveorder-2.2.0/waveorder/models/inplane_oriented_thick_pol3d_vector.py +0 -351
- waveorder-2.2.0/waveorder/sampling.py +0 -94
- waveorder-2.2.0/waveorder/visuals/matplotlib_visuals.py +0 -335
- waveorder-2.2.0/waveorder/visuals/napari_visuals.py +0 -77
- waveorder-2.2.0/waveorder/visuals/utils.py +0 -31
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/.git-blame-ignore-revs +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/.github/workflows/pytests.yml +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/CITATION.cff +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/LICENSE +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/docs/valuable-prs/2023-02-27.110.pr.open.md +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/documentation/PTI_experiment/PTI_Experiment_Recon3D_anisotropic_target_small.pdf +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/documentation/PTI_experiment/PTI_full_FOV_anisotropic_target.ipynb +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/documentation/PTI_experiment/PTI_full_FOV_cardiac_muscle.ipynb +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/documentation/PTI_experiment/PTI_full_FOV_cardiomyocyte_infected_1.ipynb +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/documentation/PTI_experiment/PTI_full_FOV_cardiomyocyte_infected_2.ipynb +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/documentation/PTI_experiment/PTI_full_FOV_cardiomyocyte_mock.ipynb +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/documentation/PTI_experiment/PTI_full_FOV_human_uterus.ipynb +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/documentation/PTI_experiment/PTI_full_FOV_mouse_brain_aco.ipynb +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/documentation/PTI_experiment/README.md +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/documentation/QLIPP_experiment/2D_QLIPP_recon_experiment.ipynb +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/documentation/QLIPP_experiment/3D_QLIPP_recon_experiment.ipynb +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/documentation/README.md +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/documentation/fluorescence_deconvolution/fluorescence_deconv.ipynb +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/maintenance/PTI_simulation/PTI_formulation.html +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/maintenance/PTI_simulation/README.md +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/maintenance/README.md +0 -0
- /waveorder-2.2.0/examples/models/inplane_oriented_thick_pol3d.py → /waveorder-2.2.0rc0/examples/models/inplane_oriented_thick_pol3D.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/models/isotropic_fluorescent_thick_3d.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/models/isotropic_thin_3d.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/models/phase_thick_3d.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/readme.png +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/setup.cfg +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/tests/__init__.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/tests/conftest.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/tests/models/test_inplane_oriented_thick_pol3D.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/tests/models/test_isotropic_fluorescent_thick_3d.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/tests/models/test_isotropic_thin_3d.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/tests/models/test_phase_thick_3d.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/tests/test_correction.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/tests/test_examples.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/tests/test_focus_estimator.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/tests/test_stokes.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder/__init__.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder/background_estimator.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder/correction.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder/focus.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder/stokes.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder/waveorder_simulator.py +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder.egg-info/dependency_links.txt +0 -0
- {waveorder-2.2.0 → waveorder-2.2.0rc0}/waveorder.egg-info/top_level.txt +0 -0
|
@@ -1,6 +1,6 @@
|
|
|
1
1
|
Metadata-Version: 2.1
|
|
2
2
|
Name: waveorder
|
|
3
|
-
Version: 2.2.
|
|
3
|
+
Version: 2.2.0rc0
|
|
4
4
|
Summary: Wave-optical simulations and deconvolution of optical properties
|
|
5
5
|
Author-email: CZ Biohub SF <compmicro@czbiohub.org>
|
|
6
6
|
Maintainer-email: Talon Chandler <talon.chandler@czbiohub.org>, Shalin Mehta <shalin.mehta@czbiohub.org>
|
|
@@ -53,99 +53,59 @@ Classifier: Operating System :: MacOS
|
|
|
53
53
|
Requires-Python: >=3.10
|
|
54
54
|
Description-Content-Type: text/markdown
|
|
55
55
|
License-File: LICENSE
|
|
56
|
-
Requires-Dist: numpy
|
|
56
|
+
Requires-Dist: numpy<2,>=1.21
|
|
57
57
|
Requires-Dist: matplotlib>=3.1.1
|
|
58
58
|
Requires-Dist: scipy>=1.3.0
|
|
59
59
|
Requires-Dist: pywavelets>=1.1.1
|
|
60
60
|
Requires-Dist: ipywidgets>=7.5.1
|
|
61
|
-
Requires-Dist: torch>=2.
|
|
61
|
+
Requires-Dist: torch>=2.2.1
|
|
62
62
|
Provides-Extra: dev
|
|
63
63
|
Requires-Dist: pytest; extra == "dev"
|
|
64
64
|
Requires-Dist: pytest-cov; extra == "dev"
|
|
65
|
-
Provides-Extra: examples
|
|
66
|
-
Requires-Dist: napari[all]; extra == "examples"
|
|
67
|
-
Requires-Dist: jupyter; extra == "examples"
|
|
68
65
|
|
|
69
66
|
# waveorder
|
|
70
67
|
|
|
71
|
-
[](https://pypi.org/project/waveorder)
|
|
72
|
-
[](https://pypistats.org/packages/waveorder)
|
|
73
|
-
[](https://pepy.tech/project/waveorder)
|
|
74
|
-
[](https://github.com/mehta-lab/waveorder/graphs/contributors)
|
|
75
|
-

|
|
76
|
-

|
|
77
68
|

|
|
78
|
-
|
|
69
|
+
[](https://pepy.tech/project/waveorder)
|
|
70
|
+
[](https://pypi.org/project/waveorder)
|
|
71
|
+
[](https://en.wikipedia.org/wiki/Software_release_life_cycle#Alpha)
|
|
79
72
|
|
|
80
73
|
This computational imaging library enables wave-optical simulation and reconstruction of optical properties that report microscopic architectural order.
|
|
81
74
|
|
|
82
|
-
## Computational label-
|
|
75
|
+
## Computational label-free imaging
|
|
83
76
|
|
|
84
|
-
|
|
77
|
+
This vectorial wave simulator and reconstructor enabled the development of a new label-free imaging method, __permittivity tensor imaging (PTI)__, that measures density and 3D orientation of biomolecules with diffraction-limited resolution. These measurements are reconstructed from polarization-resolved images acquired with a sequence of oblique illuminations.
|
|
85
78
|
|
|
86
|
-
|
|
87
|
-
<details>
|
|
88
|
-
<summary> Chandler et al. 2024 </summary>
|
|
89
|
-
<pre><code>
|
|
90
|
-
@article{chandler_2024,
|
|
91
|
-
author = {Chandler, Talon and Hirata-Miyasaki, Eduardo and Ivanov, Ivan E. and Liu, Ziwen and Sundarraman, Deepika and Ryan, Allyson Quinn and Jacobo, Adrian and Balla, Keir and Mehta, Shalin B.},
|
|
92
|
-
title = {waveOrder: generalist framework for label-agnostic computational microscopy},
|
|
93
|
-
journal = {arXiv},
|
|
94
|
-
year = {2024},
|
|
95
|
-
month = dec,
|
|
96
|
-
eprint = {2412.09775},
|
|
97
|
-
doi = {10.48550/arXiv.2412.09775}
|
|
98
|
-
}
|
|
99
|
-
</code></pre>
|
|
100
|
-
</details>
|
|
79
|
+
The acquisition, calibration, background correction, reconstruction, and applications of PTI are described in the following [preprint](https://doi.org/10.1101/2020.12.15.422951):
|
|
101
80
|
|
|
102
|
-
|
|
81
|
+
```bibtex
|
|
82
|
+
L.-H. Yeh, I. E. Ivanov, B. B. Chhun, S.-M. Guo, E. Hashemi, J. R. Byrum, J. A. Pérez-Bermejo, H. Wang, Y. Yu, P. G. Kazansky, B. R. Conklin, M. H. Han, and S. B. Mehta, "uPTI: uniaxial permittivity tensor imaging of intrinsic density and anisotropy," bioRxiv 2020.12.15.422951 (2020).
|
|
83
|
+
```
|
|
103
84
|
|
|
104
|
-
|
|
85
|
+
In addition to PTI, `waveorder` enables simulations and reconstructions of subsets of label-free measurements with subsets of the acquired data:
|
|
105
86
|
|
|
106
|
-
|
|
87
|
+
1. Reconstruction of 2D or 3D phase, projected retardance, and in-plane orientation from a polarization-diverse volumetric brightfield acquisition ([QLIPP](https://elifesciences.org/articles/55502))
|
|
107
88
|
|
|
108
|
-
|
|
89
|
+
2. Reconstruction of 2D or 3D phase from a volumetric brightfield acquisition ([2D](https://www.osapublishing.org/ao/abstract.cfm?uri=ao-54-28-8566)/[3D (PODT)](https://www.osapublishing.org/ao/abstract.cfm?uri=ao-57-1-a205) phase)
|
|
109
90
|
|
|
110
|
-
|
|
91
|
+
3. Reconstruction of 2D or 3D phase from an illumination-diverse volumetric acquisition ([2D](https://www.osapublishing.org/oe/fulltext.cfm?uri=oe-23-9-11394&id=315599)/[3D](https://www.osapublishing.org/boe/fulltext.cfm?uri=boe-7-10-3940&id=349951) differential phase contrast)
|
|
111
92
|
|
|
112
|
-
|
|
113
|
-
|
|
114
|
-
If you are interested in deploying QLIPP, phase from brightfield, or fluorescence deconvolution for label-agnostic imaging at scale, checkout our [napari plugin](https://www.napari-hub.org/plugins/recOrder-napari), [`recOrder-napari`](https://github.com/mehta-lab/recOrder).
|
|
93
|
+
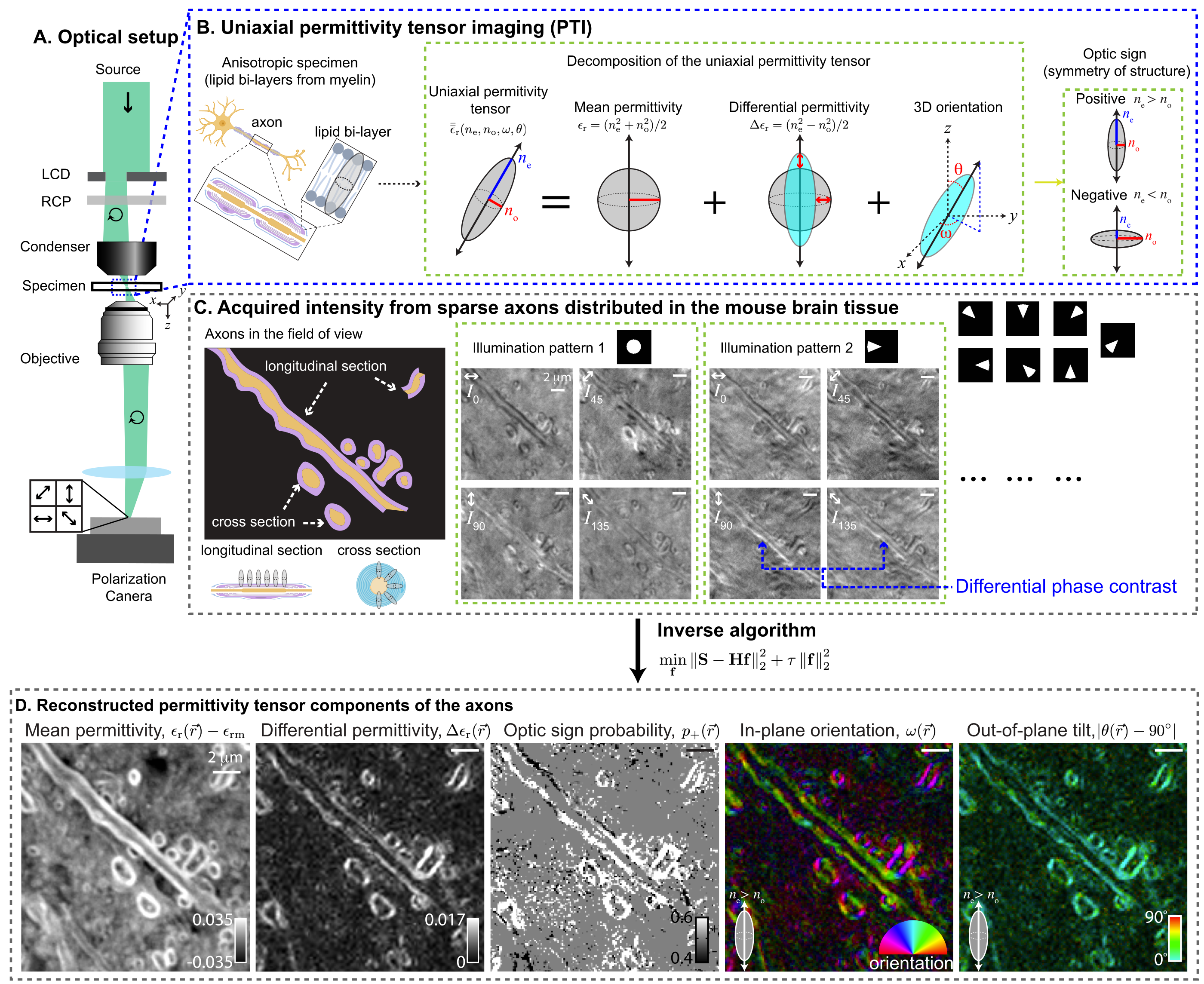
PTI provides volumetric reconstructions of mean permittivity ($\propto$ material density), differential permittivity ($\propto$ material anisotropy), 3D orientation, and optic sign. The following figure summarizes PTI acquisition and reconstruction with a small optical section of the mouse brain tissue:
|
|
115
94
|
|
|
116
|
-
|
|
95
|
+
|
|
117
96
|
|
|
118
|
-
Additionally, `waveorder` enabled the development of a new label-free imaging method, __permittivity tensor imaging (PTI)__, that measures density and 3D orientation of biomolecules with diffraction-limited resolution. These measurements are reconstructed from polarization-resolved images acquired with a sequence of oblique illuminations.
|
|
119
97
|
|
|
120
|
-
The
|
|
98
|
+
The [examples](https://github.com/mehta-lab/waveorder/tree/main/examples) illustrate simulations and reconstruction for 2D QLIPP, 3D PODT, and 2D/3D PTI methods.
|
|
121
99
|
|
|
122
|
-
|
|
123
|
-
<summary> Yeh et al. 2024 </summary>
|
|
124
|
-
<pre><code>
|
|
125
|
-
@article{yeh_2024,
|
|
126
|
-
author = {Yeh, Li-Hao and Ivanov, Ivan E. and Chandler, Talon and Byrum, Janie R. and Chhun, Bryant B. and Guo, Syuan-Ming and Foltz, Cameron and Hashemi, Ezzat and Perez-Bermejo, Juan A. and Wang, Huijun and Yu, Yanhao and Kazansky, Peter G. and Conklin, Bruce R. and Han, May H. and Mehta, Shalin B.},
|
|
127
|
-
title = {Permittivity tensor imaging: modular label-free imaging of 3D dry mass and 3D orientation at high resolution},
|
|
128
|
-
journal = {Nature Methods},
|
|
129
|
-
volume = {21},
|
|
130
|
-
number = {7},
|
|
131
|
-
pages = {1257--1274},
|
|
132
|
-
year = {2024},
|
|
133
|
-
month = jul,
|
|
134
|
-
issn = {1548-7105},
|
|
135
|
-
publisher = {Nature Publishing Group},
|
|
136
|
-
doi = {10.1038/s41592-024-02291-w}
|
|
137
|
-
}
|
|
138
|
-
</code></pre>
|
|
139
|
-
</details>
|
|
100
|
+
If you are interested in deploying QLIPP or PODT for label-free imaging at scale, checkout our [napari plugin](https://www.napari-hub.org/plugins/recOrder-napari), [`recOrder-napari`](https://github.com/mehta-lab/recOrder).
|
|
140
101
|
|
|
141
|
-
|
|
102
|
+
## Correlative imaging
|
|
142
103
|
|
|
143
|
-
|
|
104
|
+
In addition to label-free reconstruction algorithms, `waveorder` also implements widefield fluorescence and fluorescence polarization reconstruction algorithms for correlative label-free and fluorescence imaging.
|
|
144
105
|
|
|
145
|
-
|
|
146
|
-
The [examples](https://github.com/mehta-lab/waveorder/tree/main/examples) illustrate simulations and reconstruction for 2D QLIPP, 3D phase from brightfield, and 2D/3D PTI methods.
|
|
106
|
+
1. Correlative measurements of biomolecular density and orientation from polarization-diverse widefield imaging ([multimodal Instant PolScope](https://opg.optica.org/boe/fulltext.cfm?uri=boe-13-5-3102&id=472350))
|
|
147
107
|
|
|
148
|
-
|
|
108
|
+
We provide an [example notebook](https://github.com/mehta-lab/waveorder/blob/main/examples/documentation/fluorescence_deconvolution/fluorescence_deconv.ipynb) for widefield fluorescence deconvolution.
|
|
149
109
|
|
|
150
110
|
## Citation
|
|
151
111
|
|
|
@@ -153,10 +113,10 @@ Please cite this repository, along with the relevant preprint or paper, if you u
|
|
|
153
113
|
|
|
154
114
|
## Installation
|
|
155
115
|
|
|
156
|
-
|
|
116
|
+
(Optional but recommended) install [anaconda](https://www.anaconda.com/products/distribution) and create a virtual environment:
|
|
157
117
|
|
|
158
118
|
```sh
|
|
159
|
-
conda create -y -n waveorder python=3.
|
|
119
|
+
conda create -y -n waveorder python=3.11
|
|
160
120
|
conda activate waveorder
|
|
161
121
|
```
|
|
162
122
|
|
|
@@ -173,11 +133,12 @@ python
|
|
|
173
133
|
>>> import waveorder
|
|
174
134
|
```
|
|
175
135
|
|
|
176
|
-
(Optional)
|
|
136
|
+
(Optional) Download the repository, install `jupyter`, and experiment with the example notebooks
|
|
137
|
+
|
|
177
138
|
```sh
|
|
178
|
-
pip install waveorder[examples]
|
|
179
139
|
git clone https://github.com/mehta-lab/waveorder.git
|
|
180
|
-
|
|
140
|
+
pip install jupyter
|
|
141
|
+
jupyter notebook ./waveorder/examples/
|
|
181
142
|
```
|
|
182
143
|
|
|
183
144
|
(M1 users) `pytorch` has [incomplete GPU support](https://github.com/pytorch/pytorch/issues/77764),
|
|
@@ -0,0 +1,82 @@
|
|
|
1
|
+
# waveorder
|
|
2
|
+
|
|
3
|
+

|
|
4
|
+
[](https://pepy.tech/project/waveorder)
|
|
5
|
+
[](https://pypi.org/project/waveorder)
|
|
6
|
+
[](https://en.wikipedia.org/wiki/Software_release_life_cycle#Alpha)
|
|
7
|
+
|
|
8
|
+
This computational imaging library enables wave-optical simulation and reconstruction of optical properties that report microscopic architectural order.
|
|
9
|
+
|
|
10
|
+
## Computational label-free imaging
|
|
11
|
+
|
|
12
|
+
This vectorial wave simulator and reconstructor enabled the development of a new label-free imaging method, __permittivity tensor imaging (PTI)__, that measures density and 3D orientation of biomolecules with diffraction-limited resolution. These measurements are reconstructed from polarization-resolved images acquired with a sequence of oblique illuminations.
|
|
13
|
+
|
|
14
|
+
The acquisition, calibration, background correction, reconstruction, and applications of PTI are described in the following [preprint](https://doi.org/10.1101/2020.12.15.422951):
|
|
15
|
+
|
|
16
|
+
```bibtex
|
|
17
|
+
L.-H. Yeh, I. E. Ivanov, B. B. Chhun, S.-M. Guo, E. Hashemi, J. R. Byrum, J. A. Pérez-Bermejo, H. Wang, Y. Yu, P. G. Kazansky, B. R. Conklin, M. H. Han, and S. B. Mehta, "uPTI: uniaxial permittivity tensor imaging of intrinsic density and anisotropy," bioRxiv 2020.12.15.422951 (2020).
|
|
18
|
+
```
|
|
19
|
+
|
|
20
|
+
In addition to PTI, `waveorder` enables simulations and reconstructions of subsets of label-free measurements with subsets of the acquired data:
|
|
21
|
+
|
|
22
|
+
1. Reconstruction of 2D or 3D phase, projected retardance, and in-plane orientation from a polarization-diverse volumetric brightfield acquisition ([QLIPP](https://elifesciences.org/articles/55502))
|
|
23
|
+
|
|
24
|
+
2. Reconstruction of 2D or 3D phase from a volumetric brightfield acquisition ([2D](https://www.osapublishing.org/ao/abstract.cfm?uri=ao-54-28-8566)/[3D (PODT)](https://www.osapublishing.org/ao/abstract.cfm?uri=ao-57-1-a205) phase)
|
|
25
|
+
|
|
26
|
+
3. Reconstruction of 2D or 3D phase from an illumination-diverse volumetric acquisition ([2D](https://www.osapublishing.org/oe/fulltext.cfm?uri=oe-23-9-11394&id=315599)/[3D](https://www.osapublishing.org/boe/fulltext.cfm?uri=boe-7-10-3940&id=349951) differential phase contrast)
|
|
27
|
+
|
|
28
|
+
PTI provides volumetric reconstructions of mean permittivity ($\propto$ material density), differential permittivity ($\propto$ material anisotropy), 3D orientation, and optic sign. The following figure summarizes PTI acquisition and reconstruction with a small optical section of the mouse brain tissue:
|
|
29
|
+
|
|
30
|
+

|
|
31
|
+
|
|
32
|
+
|
|
33
|
+
The [examples](https://github.com/mehta-lab/waveorder/tree/main/examples) illustrate simulations and reconstruction for 2D QLIPP, 3D PODT, and 2D/3D PTI methods.
|
|
34
|
+
|
|
35
|
+
If you are interested in deploying QLIPP or PODT for label-free imaging at scale, checkout our [napari plugin](https://www.napari-hub.org/plugins/recOrder-napari), [`recOrder-napari`](https://github.com/mehta-lab/recOrder).
|
|
36
|
+
|
|
37
|
+
## Correlative imaging
|
|
38
|
+
|
|
39
|
+
In addition to label-free reconstruction algorithms, `waveorder` also implements widefield fluorescence and fluorescence polarization reconstruction algorithms for correlative label-free and fluorescence imaging.
|
|
40
|
+
|
|
41
|
+
1. Correlative measurements of biomolecular density and orientation from polarization-diverse widefield imaging ([multimodal Instant PolScope](https://opg.optica.org/boe/fulltext.cfm?uri=boe-13-5-3102&id=472350))
|
|
42
|
+
|
|
43
|
+
We provide an [example notebook](https://github.com/mehta-lab/waveorder/blob/main/examples/documentation/fluorescence_deconvolution/fluorescence_deconv.ipynb) for widefield fluorescence deconvolution.
|
|
44
|
+
|
|
45
|
+
## Citation
|
|
46
|
+
|
|
47
|
+
Please cite this repository, along with the relevant preprint or paper, if you use or adapt this code. The citation information can be found by clicking "Cite this repository" button in the About section in the right sidebar.
|
|
48
|
+
|
|
49
|
+
## Installation
|
|
50
|
+
|
|
51
|
+
(Optional but recommended) install [anaconda](https://www.anaconda.com/products/distribution) and create a virtual environment:
|
|
52
|
+
|
|
53
|
+
```sh
|
|
54
|
+
conda create -y -n waveorder python=3.11
|
|
55
|
+
conda activate waveorder
|
|
56
|
+
```
|
|
57
|
+
|
|
58
|
+
Install `waveorder` from PyPI:
|
|
59
|
+
|
|
60
|
+
```sh
|
|
61
|
+
pip install waveorder
|
|
62
|
+
```
|
|
63
|
+
|
|
64
|
+
Use `waveorder` in your scripts:
|
|
65
|
+
|
|
66
|
+
```sh
|
|
67
|
+
python
|
|
68
|
+
>>> import waveorder
|
|
69
|
+
```
|
|
70
|
+
|
|
71
|
+
(Optional) Download the repository, install `jupyter`, and experiment with the example notebooks
|
|
72
|
+
|
|
73
|
+
```sh
|
|
74
|
+
git clone https://github.com/mehta-lab/waveorder.git
|
|
75
|
+
pip install jupyter
|
|
76
|
+
jupyter notebook ./waveorder/examples/
|
|
77
|
+
```
|
|
78
|
+
|
|
79
|
+
(M1 users) `pytorch` has [incomplete GPU support](https://github.com/pytorch/pytorch/issues/77764),
|
|
80
|
+
so please use `export PYTORCH_ENABLE_MPS_FALLBACK=1`
|
|
81
|
+
to allow some operators to fallback to CPU
|
|
82
|
+
if you plan to use GPU acceleration for polarization reconstruction.
|
|
@@ -0,0 +1,7 @@
|
|
|
1
|
+
`waveorder` is undergoing a significant refactor, and this `examples/` folder serves as a good place to understand the current state of the repository.
|
|
2
|
+
|
|
3
|
+
The `models/` folder demonstrates the latest functionality of `waveorder`. These scripts will run as is in an environment with `waveorder` and `napari` installed. Each script demonstrates a simulation and reconstruction with a **model**: a specific set of assumptions about the sample and the data being acquired.
|
|
4
|
+
|
|
5
|
+
The `maintenance/` folder demonstrates the functionality of `waveorder` that we plan to move to `models/`. These scripts can be run as is, and they are being maintained with tests.
|
|
6
|
+
|
|
7
|
+
The `documentation/` folder consists of examples that demonstrate reconstruction with real data. These examples require access to the complete datasets, so they are not being actively maintained and serve primarily as documentation.
|
|
@@ -6,12 +6,11 @@ import matplotlib.pyplot as plt
|
|
|
6
6
|
from numpy.fft import fftshift
|
|
7
7
|
|
|
8
8
|
import waveorder as wo
|
|
9
|
-
from waveorder import optics, waveorder_reconstructor, util
|
|
9
|
+
from waveorder import optics, waveorder_reconstructor, util, visual
|
|
10
10
|
|
|
11
11
|
import zarr
|
|
12
12
|
from pathlib import Path
|
|
13
13
|
from iohub import open_ome_zarr
|
|
14
|
-
from waveorder.visuals import jupyter_visuals
|
|
15
14
|
|
|
16
15
|
# %%
|
|
17
16
|
# Initialization
|
|
@@ -111,7 +110,7 @@ for i in range(len(Source)):
|
|
|
111
110
|
Source_PolState[i, 1] = E_in[1]
|
|
112
111
|
|
|
113
112
|
|
|
114
|
-
|
|
113
|
+
visual.plot_multicolumn(
|
|
115
114
|
fftshift(Source, axes=(1, 2)), origin="lower", num_col=5
|
|
116
115
|
)
|
|
117
116
|
|
|
@@ -163,7 +162,7 @@ S_image_tm[2] = (
|
|
|
163
162
|
|
|
164
163
|
# %%
|
|
165
164
|
# browse raw intensity stacks (stack_idx_1: z index, stack_idx2: pattern index)
|
|
166
|
-
|
|
165
|
+
visual.parallel_5D_viewer(
|
|
167
166
|
np.transpose(I_meas[:, :, :, :, ::-1], (4, 1, 0, 2, 3)),
|
|
168
167
|
num_col=4,
|
|
169
168
|
size=10,
|
|
@@ -172,7 +171,7 @@ jupyter_visuals.parallel_5D_viewer(
|
|
|
172
171
|
|
|
173
172
|
# %%
|
|
174
173
|
# browse uncorrected Stokes parameters (stack_idx_1: z index, stack_idx2: pattern index)
|
|
175
|
-
|
|
174
|
+
visual.parallel_5D_viewer(
|
|
176
175
|
np.transpose(S_image_recon, (4, 1, 0, 2, 3)),
|
|
177
176
|
num_col=3,
|
|
178
177
|
size=8,
|
|
@@ -183,7 +182,7 @@ jupyter_visuals.parallel_5D_viewer(
|
|
|
183
182
|
|
|
184
183
|
# %%
|
|
185
184
|
# browse corrected Stokes parameters (stack_idx_1: z index, stack_idx2: pattern index)
|
|
186
|
-
|
|
185
|
+
visual.parallel_5D_viewer(
|
|
187
186
|
np.transpose(S_image_tm, (4, 1, 0, 2, 3)),
|
|
188
187
|
num_col=3,
|
|
189
188
|
size=8,
|
|
@@ -214,7 +213,7 @@ f_tensor = setup.scattering_potential_tensor_recon_3D_vec(
|
|
|
214
213
|
|
|
215
214
|
# %%
|
|
216
215
|
# browse the z-stack of components of scattering potential tensor
|
|
217
|
-
|
|
216
|
+
visual.parallel_4D_viewer(
|
|
218
217
|
np.transpose(f_tensor, (3, 0, 1, 2)),
|
|
219
218
|
num_col=4,
|
|
220
219
|
origin="lower",
|
|
@@ -279,7 +278,7 @@ differential_permittivity_PT = np.array(
|
|
|
279
278
|
|
|
280
279
|
# %%
|
|
281
280
|
# browse the reconstructed physical properties
|
|
282
|
-
|
|
281
|
+
visual.parallel_4D_viewer(
|
|
283
282
|
np.transpose(
|
|
284
283
|
np.stack(
|
|
285
284
|
[
|
|
@@ -547,7 +546,7 @@ ax[5, 1].set_title("inclination (+) (xz)")
|
|
|
547
546
|
|
|
548
547
|
# %%
|
|
549
548
|
# browse XY planes of the phase and differential permittivity
|
|
550
|
-
|
|
549
|
+
visual.parallel_4D_viewer(
|
|
551
550
|
np.transpose(
|
|
552
551
|
[
|
|
553
552
|
np.clip(phase_PT, phase_min, phase_max),
|
|
@@ -586,7 +585,7 @@ orientation_3D_image = np.transpose(
|
|
|
586
585
|
),
|
|
587
586
|
(3, 1, 2, 0),
|
|
588
587
|
)
|
|
589
|
-
orientation_3D_image_RGB =
|
|
588
|
+
orientation_3D_image_RGB = visual.orientation_3D_to_rgb(
|
|
590
589
|
orientation_3D_image, interp_belt=20 / 180 * np.pi, sat_factor=1
|
|
591
590
|
)
|
|
592
591
|
|
|
@@ -601,7 +600,7 @@ plt.imshow(
|
|
|
601
600
|
|
|
602
601
|
# plot the top view of 3D orientation colorsphere
|
|
603
602
|
plt.figure(figsize=(3, 3))
|
|
604
|
-
|
|
603
|
+
visual.orientation_3D_colorwheel(
|
|
605
604
|
wheelsize=256, circ_size=50, interp_belt=20 / 180 * np.pi, sat_factor=1
|
|
606
605
|
)
|
|
607
606
|
|
|
@@ -640,7 +639,7 @@ plt.imshow(
|
|
|
640
639
|
in_plane_orientation[:, y_layer], origin="lower", aspect=z_step / ps
|
|
641
640
|
)
|
|
642
641
|
plt.figure(figsize=(3, 3))
|
|
643
|
-
|
|
642
|
+
visual.orientation_2D_colorwheel()
|
|
644
643
|
|
|
645
644
|
# %%
|
|
646
645
|
# out-of-plane tilt
|
|
@@ -687,7 +686,7 @@ z_layer = 44
|
|
|
687
686
|
|
|
688
687
|
fig, ax = plt.subplots(1, 1, figsize=(15, 15))
|
|
689
688
|
|
|
690
|
-
|
|
689
|
+
visual.plot3DVectorField(
|
|
691
690
|
np.abs(differential_permittivity_PT[1, :, :, z_layer]),
|
|
692
691
|
azimuth[1, :, :, z_layer],
|
|
693
692
|
theta[1, :, :, z_layer],
|
|
@@ -723,7 +722,7 @@ plt.imshow(
|
|
|
723
722
|
# %%
|
|
724
723
|
# Angular histogram of 3D orientation
|
|
725
724
|
|
|
726
|
-
|
|
725
|
+
visual.orientation_3D_hist(
|
|
727
726
|
azimuth[1].flatten(),
|
|
728
727
|
theta[1].flatten(),
|
|
729
728
|
ret_mask.flatten(),
|
|
@@ -15,9 +15,9 @@ from numpy.fft import fftshift
|
|
|
15
15
|
from waveorder import (
|
|
16
16
|
optics,
|
|
17
17
|
waveorder_simulator,
|
|
18
|
+
visual,
|
|
18
19
|
util,
|
|
19
20
|
)
|
|
20
|
-
from waveorder.visuals import jupyter_visuals
|
|
21
21
|
|
|
22
22
|
#####################################################################
|
|
23
23
|
# Initialization - imaging system and sample #
|
|
@@ -145,7 +145,7 @@ biref_map = ne_map_copy - no_map_copy
|
|
|
145
145
|
### Visualize sample properties
|
|
146
146
|
|
|
147
147
|
#### XY sections
|
|
148
|
-
|
|
148
|
+
visual.plot_multicolumn(
|
|
149
149
|
[
|
|
150
150
|
target[:, :, z_layer],
|
|
151
151
|
azimuth[:, :, z_layer] % (2 * np.pi),
|
|
@@ -158,7 +158,7 @@ jupyter_visuals.plot_multicolumn(
|
|
|
158
158
|
set_title=True,
|
|
159
159
|
)
|
|
160
160
|
#### XZ sections
|
|
161
|
-
|
|
161
|
+
visual.plot_multicolumn(
|
|
162
162
|
[
|
|
163
163
|
np.transpose(target[y_layer, :, :]),
|
|
164
164
|
np.transpose(azimuth[y_layer, :, :]) % (2 * np.pi),
|
|
@@ -197,7 +197,7 @@ orientation_3D_image = np.transpose(
|
|
|
197
197
|
),
|
|
198
198
|
(3, 1, 2, 0),
|
|
199
199
|
)
|
|
200
|
-
orientation_3D_image_RGB =
|
|
200
|
+
orientation_3D_image_RGB = visual.orientation_3D_to_rgb(
|
|
201
201
|
orientation_3D_image, interp_belt=20 / 180 * np.pi, sat_factor=1
|
|
202
202
|
)
|
|
203
203
|
|
|
@@ -206,7 +206,7 @@ plt.imshow(orientation_3D_image_RGB[z_layer], origin="lower")
|
|
|
206
206
|
plt.figure(figsize=(10, 10))
|
|
207
207
|
plt.imshow(orientation_3D_image_RGB[:, y_layer], origin="lower")
|
|
208
208
|
plt.figure(figsize=(3, 3))
|
|
209
|
-
|
|
209
|
+
visual.orientation_3D_colorwheel(
|
|
210
210
|
wheelsize=128,
|
|
211
211
|
circ_size=50,
|
|
212
212
|
interp_belt=20 / 180 * np.pi,
|
|
@@ -216,7 +216,7 @@ jupyter_visuals.orientation_3D_colorwheel(
|
|
|
216
216
|
plt.show()
|
|
217
217
|
|
|
218
218
|
#### Angular histogram of 3D orientation
|
|
219
|
-
|
|
219
|
+
visual.orientation_3D_hist(
|
|
220
220
|
azimuth.flatten(),
|
|
221
221
|
inclination.flatten(),
|
|
222
222
|
np.abs(target).flatten(),
|
|
@@ -258,7 +258,7 @@ epsilon_tensor[2, 1] = epsilon_del * np.sin(2 * inclination) * np.sin(azimuth)
|
|
|
258
258
|
epsilon_tensor[2, 2] = epsilon_mean + epsilon_del * np.cos(2 * inclination)
|
|
259
259
|
|
|
260
260
|
|
|
261
|
-
|
|
261
|
+
visual.plot_multicolumn(
|
|
262
262
|
[
|
|
263
263
|
epsilon_tensor[0, 0, :, :, z_layer],
|
|
264
264
|
epsilon_tensor[0, 1, :, :, z_layer],
|
|
@@ -334,7 +334,7 @@ del_f_component[6] = (
|
|
|
334
334
|
)
|
|
335
335
|
|
|
336
336
|
|
|
337
|
-
|
|
337
|
+
visual.plot_multicolumn(
|
|
338
338
|
[
|
|
339
339
|
del_f_component[0, :, :, z_layer],
|
|
340
340
|
del_f_component[1, :, :, z_layer],
|
|
@@ -425,11 +425,11 @@ for i in range(len(Source)):
|
|
|
425
425
|
|
|
426
426
|
#### Circularly polarized illumination patterns
|
|
427
427
|
|
|
428
|
-
|
|
428
|
+
visual.plot_multicolumn(
|
|
429
429
|
fftshift(Source_cont, axes=(1, 2)), origin="lower", num_col=5, size=5
|
|
430
430
|
)
|
|
431
431
|
# discretized illumination patterns used in simulation (faster forward model)
|
|
432
|
-
|
|
432
|
+
visual.plot_multicolumn(
|
|
433
433
|
fftshift(Source, axes=(1, 2)), origin="lower", num_col=5, size=5
|
|
434
434
|
)
|
|
435
435
|
print(Source_PolState)
|
{waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/maintenance/PTI_simulation/PTI_Simulation_Recon2D.py
RENAMED
|
@@ -16,8 +16,8 @@ from numpy.fft import fftshift
|
|
|
16
16
|
from waveorder import (
|
|
17
17
|
optics,
|
|
18
18
|
waveorder_reconstructor,
|
|
19
|
+
visual,
|
|
19
20
|
)
|
|
20
|
-
from waveorder.visuals import jupyter_visuals
|
|
21
21
|
|
|
22
22
|
## Initialization
|
|
23
23
|
## Load simulated images and parameters
|
|
@@ -76,7 +76,7 @@ setup = waveorder_reconstructor.waveorder_microscopy(
|
|
|
76
76
|
## Visualize 2 D transfer functions as a function of illumination pattern
|
|
77
77
|
|
|
78
78
|
# illumination patterns used
|
|
79
|
-
|
|
79
|
+
visual.plot_multicolumn(
|
|
80
80
|
fftshift(Source_cont, axes=(1, 2)), origin="lower", num_col=5, size=5
|
|
81
81
|
)
|
|
82
82
|
plt.show()
|
|
@@ -118,7 +118,7 @@ f_tensor = setup.scattering_potential_tensor_recon_2D_vec(
|
|
|
118
118
|
S_image_tm, reg_inc=reg_inc, cupy_det=True
|
|
119
119
|
)
|
|
120
120
|
|
|
121
|
-
|
|
121
|
+
visual.plot_multicolumn(
|
|
122
122
|
f_tensor,
|
|
123
123
|
num_col=4,
|
|
124
124
|
origin="lower",
|
|
@@ -255,14 +255,14 @@ orientation_3D_image = np.transpose(
|
|
|
255
255
|
),
|
|
256
256
|
(1, 2, 0),
|
|
257
257
|
)
|
|
258
|
-
orientation_3D_image_RGB =
|
|
258
|
+
orientation_3D_image_RGB = visual.orientation_3D_to_rgb(
|
|
259
259
|
orientation_3D_image, interp_belt=20 / 180 * np.pi, sat_factor=1
|
|
260
260
|
)
|
|
261
261
|
|
|
262
262
|
plt.figure(figsize=(5, 5))
|
|
263
263
|
plt.imshow(orientation_3D_image_RGB, origin="lower")
|
|
264
264
|
plt.figure(figsize=(3, 3))
|
|
265
|
-
|
|
265
|
+
visual.orientation_3D_colorwheel(
|
|
266
266
|
wheelsize=256, circ_size=50, interp_belt=20 / 180 * np.pi, sat_factor=1
|
|
267
267
|
)
|
|
268
268
|
plt.show()
|
|
@@ -297,7 +297,7 @@ in_plane_orientation = hsv_to_rgb(I_hsv.copy())
|
|
|
297
297
|
plt.figure(figsize=(5, 5))
|
|
298
298
|
plt.imshow(in_plane_orientation, origin="lower")
|
|
299
299
|
plt.figure(figsize=(3, 3))
|
|
300
|
-
|
|
300
|
+
visual.orientation_2D_colorwheel()
|
|
301
301
|
plt.show()
|
|
302
302
|
|
|
303
303
|
# out-of-plane tilt
|
|
@@ -339,7 +339,7 @@ spacing = 4
|
|
|
339
339
|
plt.figure(figsize=(10, 10))
|
|
340
340
|
|
|
341
341
|
fig, ax = plt.subplots(1, 1, figsize=(20, 10))
|
|
342
|
-
|
|
342
|
+
visual.plot3DVectorField(
|
|
343
343
|
np.abs(retardance_pr_nm[0]),
|
|
344
344
|
azimuth[0],
|
|
345
345
|
theta[0],
|
|
@@ -362,7 +362,7 @@ ret_mask[ret_mask < 0.5] = 0
|
|
|
362
362
|
|
|
363
363
|
plt.figure(figsize=(10, 10))
|
|
364
364
|
plt.imshow(ret_mask, cmap="gray", origin="lower")
|
|
365
|
-
|
|
365
|
+
visual.orientation_3D_hist(
|
|
366
366
|
azimuth[0].flatten(),
|
|
367
367
|
theta[0].flatten(),
|
|
368
368
|
ret_mask.flatten(),
|
{waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/maintenance/PTI_simulation/PTI_Simulation_Recon3D.py
RENAMED
|
@@ -14,8 +14,8 @@ from numpy.fft import fftshift
|
|
|
14
14
|
from waveorder import (
|
|
15
15
|
optics,
|
|
16
16
|
waveorder_reconstructor,
|
|
17
|
+
visual,
|
|
17
18
|
)
|
|
18
|
-
from waveorder.visuals import jupyter_visuals
|
|
19
19
|
|
|
20
20
|
## Initialization
|
|
21
21
|
## Load simulated images and parameters
|
|
@@ -65,7 +65,7 @@ setup = waveorder_reconstructor.waveorder_microscopy(
|
|
|
65
65
|
|
|
66
66
|
|
|
67
67
|
### Illumination patterns used
|
|
68
|
-
|
|
68
|
+
visual.plot_multicolumn(
|
|
69
69
|
fftshift(Source_cont, axes=(1, 2)), origin="lower", num_col=5, size=5
|
|
70
70
|
)
|
|
71
71
|
plt.show()
|
|
@@ -113,7 +113,7 @@ f_tensor = setup.scattering_potential_tensor_recon_3D_vec(
|
|
|
113
113
|
S_image_tm, reg_inc=reg_inc, cupy_det=True
|
|
114
114
|
)
|
|
115
115
|
|
|
116
|
-
|
|
116
|
+
visual.plot_multicolumn(
|
|
117
117
|
f_tensor[..., L // 2],
|
|
118
118
|
num_col=4,
|
|
119
119
|
origin="lower",
|
|
@@ -183,7 +183,7 @@ phase_PT, absorption_PT, retardance_pr_PT = [
|
|
|
183
183
|
### Reconstructed phase, absorption, principal retardance, azimuth, and inclination assuming (+) and (-) optic sign
|
|
184
184
|
|
|
185
185
|
# browse the reconstructed physical properties
|
|
186
|
-
|
|
186
|
+
visual.plot_multicolumn(
|
|
187
187
|
np.stack(
|
|
188
188
|
[
|
|
189
189
|
phase_PT[..., L // 2],
|
|
@@ -389,7 +389,7 @@ orientation_3D_image = np.transpose(
|
|
|
389
389
|
),
|
|
390
390
|
(3, 1, 2, 0),
|
|
391
391
|
)
|
|
392
|
-
orientation_3D_image_RGB =
|
|
392
|
+
orientation_3D_image_RGB = visual.orientation_3D_to_rgb(
|
|
393
393
|
orientation_3D_image, interp_belt=20 / 180 * np.pi, sat_factor=1
|
|
394
394
|
)
|
|
395
395
|
|
|
@@ -402,7 +402,7 @@ plt.imshow(
|
|
|
402
402
|
)
|
|
403
403
|
# plot the top view of 3D orientation colorsphere
|
|
404
404
|
plt.figure(figsize=(3, 3))
|
|
405
|
-
|
|
405
|
+
visual.orientation_3D_colorwheel(
|
|
406
406
|
wheelsize=128,
|
|
407
407
|
circ_size=50,
|
|
408
408
|
interp_belt=20 / 180 * np.pi,
|
|
@@ -440,7 +440,7 @@ plt.imshow(in_plane_orientation[z_layer], origin="lower")
|
|
|
440
440
|
plt.figure(figsize=(10, 10))
|
|
441
441
|
plt.imshow(in_plane_orientation[:, y_layer], origin="lower", aspect=psz / ps)
|
|
442
442
|
plt.figure(figsize=(3, 3))
|
|
443
|
-
|
|
443
|
+
visual.orientation_2D_colorwheel()
|
|
444
444
|
plt.show()
|
|
445
445
|
|
|
446
446
|
|
|
@@ -511,7 +511,7 @@ linelength_scale = 20
|
|
|
511
511
|
|
|
512
512
|
|
|
513
513
|
fig, ax = plt.subplots(2, 2, figsize=(10, 10))
|
|
514
|
-
|
|
514
|
+
visual.plot3DVectorField(
|
|
515
515
|
np.abs(retardance_pr_PT[0, :, :, z_layer]),
|
|
516
516
|
azimuth[0, :, :, z_layer],
|
|
517
517
|
theta[0, :, :, z_layer],
|
|
@@ -529,7 +529,7 @@ jupyter_visuals.plot3DVectorField(
|
|
|
529
529
|
)
|
|
530
530
|
ax[0, 0].set_title(f"XY section (z= {z_layer})")
|
|
531
531
|
|
|
532
|
-
|
|
532
|
+
visual.plot3DVectorField(
|
|
533
533
|
np.transpose(np.abs(retardance_pr_PT[0, :, x_layer, :])),
|
|
534
534
|
np.transpose(azimuth_x[0, :, x_layer, :]),
|
|
535
535
|
np.transpose(theta_x[0, :, x_layer, :]),
|
|
@@ -547,7 +547,7 @@ jupyter_visuals.plot3DVectorField(
|
|
|
547
547
|
)
|
|
548
548
|
ax[0, 1].set_title(f"YZ section (x = {x_layer})")
|
|
549
549
|
|
|
550
|
-
|
|
550
|
+
visual.plot3DVectorField(
|
|
551
551
|
np.transpose(np.abs(retardance_pr_PT[0, y_layer, :, :])),
|
|
552
552
|
np.transpose(azimuth_y[0, y_layer, :, :]),
|
|
553
553
|
np.transpose(theta_y[0, y_layer, :, :]),
|
|
@@ -584,7 +584,7 @@ ret_mask[ret_mask < 0.00125] = 0
|
|
|
584
584
|
|
|
585
585
|
plt.figure(figsize=(10, 10))
|
|
586
586
|
plt.imshow(ret_mask[:, :, z_layer], cmap="gray", origin="lower")
|
|
587
|
-
|
|
587
|
+
visual.orientation_3D_hist(
|
|
588
588
|
azimuth[0].flatten(),
|
|
589
589
|
theta[0].flatten(),
|
|
590
590
|
ret_mask.flatten(),
|
{waveorder-2.2.0 → waveorder-2.2.0rc0}/examples/maintenance/QLIPP_simulation/2D_QLIPP_forward.py
RENAMED
|
@@ -16,9 +16,9 @@ from numpy.fft import fftshift
|
|
|
16
16
|
from waveorder import (
|
|
17
17
|
optics,
|
|
18
18
|
waveorder_simulator,
|
|
19
|
+
visual,
|
|
19
20
|
util,
|
|
20
21
|
)
|
|
21
|
-
from waveorder.visuals import jupyter_visuals
|
|
22
22
|
|
|
23
23
|
# Key parameters
|
|
24
24
|
N = 256 # number of pixel in y dimension
|
|
@@ -38,7 +38,7 @@ chi = 0.03 * 2 * np.pi # swing of Polscope analyzer
|
|
|
38
38
|
star, theta, _ = util.generate_star_target((N, M))
|
|
39
39
|
star = star.numpy()
|
|
40
40
|
theta = theta.numpy()
|
|
41
|
-
|
|
41
|
+
visual.plot_multicolumn(np.array([star, theta]), num_col=2, size=5)
|
|
42
42
|
|
|
43
43
|
# Assign uniform phase, uniform retardance, and radial slow axes to the star pattern
|
|
44
44
|
phase_value = 1 # average phase in radians (optical path length)
|
|
@@ -50,7 +50,7 @@ t_eigen = np.zeros((2, N, M), complex) # complex specimen transmission
|
|
|
50
50
|
t_eigen[0] = np.exp(-mu_s + 1j * phi_s)
|
|
51
51
|
t_eigen[1] = np.exp(-mu_f + 1j * phi_f)
|
|
52
52
|
sa = theta % np.pi # slow axes.
|
|
53
|
-
|
|
53
|
+
visual.plot_multicolumn(
|
|
54
54
|
np.array([phi_s, phi_f, mu_s, sa]),
|
|
55
55
|
num_col=2,
|
|
56
56
|
size=5,
|