tanabesugano 1.5.0__tar.gz
This diff represents the content of publicly available package versions that have been released to one of the supported registries. The information contained in this diff is provided for informational purposes only and reflects changes between package versions as they appear in their respective public registries.
- tanabesugano-1.5.0/LICENSE +21 -0
- tanabesugano-1.5.0/PKG-INFO +150 -0
- tanabesugano-1.5.0/README.md +103 -0
- tanabesugano-1.5.0/pyproject.toml +97 -0
- tanabesugano-1.5.0/tanabesugano/__init__.py +6 -0
- tanabesugano-1.5.0/tanabesugano/batch.py +159 -0
- tanabesugano-1.5.0/tanabesugano/cmd.py +461 -0
- tanabesugano-1.5.0/tanabesugano/matrices.py +1336 -0
- tanabesugano-1.5.0/tanabesugano/test/__init__.py +0 -0
- tanabesugano-1.5.0/tanabesugano/test/test_batch.py +51 -0
- tanabesugano-1.5.0/tanabesugano/test/test_front.py +20 -0
- tanabesugano-1.5.0/tanabesugano/test/test_matrices.py +89 -0
- tanabesugano-1.5.0/tanabesugano/test/test_num.py +53 -0
- tanabesugano-1.5.0/tanabesugano/tools.py +35 -0
|
@@ -0,0 +1,21 @@
|
|
|
1
|
+
MIT License
|
|
2
|
+
|
|
3
|
+
Copyright (c) 2019 Anselm Hahn
|
|
4
|
+
|
|
5
|
+
Permission is hereby granted, free of charge, to any person obtaining a copy
|
|
6
|
+
of this software and associated documentation files (the "Software"), to deal
|
|
7
|
+
in the Software without restriction, including without limitation the rights
|
|
8
|
+
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
|
|
9
|
+
copies of the Software, and to permit persons to whom the Software is
|
|
10
|
+
furnished to do so, subject to the following conditions:
|
|
11
|
+
|
|
12
|
+
The above copyright notice and this permission notice shall be included in all
|
|
13
|
+
copies or substantial portions of the Software.
|
|
14
|
+
|
|
15
|
+
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
|
|
16
|
+
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
|
|
17
|
+
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
|
|
18
|
+
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
|
|
19
|
+
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
|
|
20
|
+
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
|
|
21
|
+
SOFTWARE.
|
|
@@ -0,0 +1,150 @@
|
|
|
1
|
+
Metadata-Version: 2.4
|
|
2
|
+
Name: tanabesugano
|
|
3
|
+
Version: 1.5.0
|
|
4
|
+
Summary: A python-solver for Tanabe-Sugano and Energy-Correlation diagrams
|
|
5
|
+
License: MIT
|
|
6
|
+
License-File: LICENSE
|
|
7
|
+
Keywords: terminal,data-visualization,tanabe-sugano,energy-correlation,complex-ions
|
|
8
|
+
Author: Anselm Hahn
|
|
9
|
+
Author-email: anselm.hahn@gmail.com
|
|
10
|
+
Maintainer: Anselm Hahn
|
|
11
|
+
Maintainer-email: anselm.hahn@gmail.com
|
|
12
|
+
Requires-Python: >=3.9,<4.0
|
|
13
|
+
Classifier: Development Status :: 5 - Production/Stable
|
|
14
|
+
Classifier: Environment :: Console
|
|
15
|
+
Classifier: Intended Audience :: Education
|
|
16
|
+
Classifier: Intended Audience :: Science/Research
|
|
17
|
+
Classifier: License :: OSI Approved :: MIT License
|
|
18
|
+
Classifier: Operating System :: MacOS
|
|
19
|
+
Classifier: Operating System :: Microsoft :: Windows
|
|
20
|
+
Classifier: Operating System :: Unix
|
|
21
|
+
Classifier: Programming Language :: Python :: 3
|
|
22
|
+
Classifier: Programming Language :: Python :: 3.9
|
|
23
|
+
Classifier: Programming Language :: Python :: 3.10
|
|
24
|
+
Classifier: Programming Language :: Python :: 3.11
|
|
25
|
+
Classifier: Programming Language :: Python :: 3.12
|
|
26
|
+
Classifier: Programming Language :: Python :: 3.13
|
|
27
|
+
Classifier: Programming Language :: Python :: 3.14
|
|
28
|
+
Classifier: Programming Language :: Python :: 3.8
|
|
29
|
+
Classifier: Topic :: Scientific/Engineering :: Chemistry
|
|
30
|
+
Classifier: Topic :: Scientific/Engineering :: Physics
|
|
31
|
+
Classifier: Topic :: Scientific/Engineering :: Visualization
|
|
32
|
+
Classifier: Topic :: System :: Shells
|
|
33
|
+
Classifier: Topic :: Utilities
|
|
34
|
+
Provides-Extra: plotly
|
|
35
|
+
Requires-Dist: matplotlib (>=3.4.2,<4.0.0)
|
|
36
|
+
Requires-Dist: numpy (>=1.24.4,<2.0.0) ; python_version == "3.8"
|
|
37
|
+
Requires-Dist: numpy (>=1.26.4,<2.0.0) ; python_version == "3.9"
|
|
38
|
+
Requires-Dist: numpy (>=2.0.2,<3.0.0) ; python_version >= "3.10" and python_version < "3.13"
|
|
39
|
+
Requires-Dist: pandas (>=1.2.4,<4.0.0)
|
|
40
|
+
Requires-Dist: plotly (>=5.13.1,<7.0.0) ; extra == "plotly"
|
|
41
|
+
Requires-Dist: prettytable (>=2.1,<4.0)
|
|
42
|
+
Requires-Dist: update (>=0.0.1,<0.0.2)
|
|
43
|
+
Project-URL: Homepage, https://pypi.org/project/TanabeSugano
|
|
44
|
+
Project-URL: Repository, https://github.com/Anselmoo/TanabeSugano
|
|
45
|
+
Description-Content-Type: text/markdown
|
|
46
|
+
|
|
47
|
+
[](https://github.com/Anselmoo/TanabeSugano/actions/workflows/python-package.yml)
|
|
48
|
+
[](https://python-poetry.org/)
|
|
49
|
+
[](https://www.codefactor.io/repository/github/anselmoo/tanabesugano)
|
|
50
|
+
[](https://zenodo.org/badge/latestdoi/206847682)
|
|
51
|
+
[](https://github.com/Anselmoo/TanabeSugano/blob/master/LICENSE)
|
|
52
|
+
[](https://github.com/Anselmoo/TanabeSugano/releases)
|
|
53
|
+
[](https://pypi.org/project/TanabeSugano/)
|
|
54
|
+
[](https://pypi.org/project/TanabeSugano/)
|
|
55
|
+
[](https://pepy.tech/project/tanabesugano)
|
|
56
|
+
[](https://colab.research.google.com/github/Anselmoo/TanabeSugano/blob/master/Tanabe_Sugano.ipynb)
|
|
57
|
+
|
|
58
|
+
|
|
59
|
+
# TanabeSugano
|
|
60
|
+
A python-based Eigensolver for Tanabe-Sugano- & Energy-Correlation-Diagrams based on the original three proposed studies of *Yukito Tanabe and Satoru Sugano* for d<sup>2</sup>-d<sup>8</sup> transition metal ions:
|
|
61
|
+
|
|
62
|
+
1. On the Absorption Spectra of Complex Ions. I
|
|
63
|
+
|
|
64
|
+
**Yukito Tanabe, Satoru Sugano**
|
|
65
|
+
*Journal of the Physical Society of Japan*, 9, 753-766 (1954)
|
|
66
|
+
**DOI:** 10.1143/JPSJ.9.753
|
|
67
|
+
https://journals.jps.jp/doi/10.1143/JPSJ.9.753
|
|
68
|
+
|
|
69
|
+
2. On the Absorption Spectra of Complex Ions II
|
|
70
|
+
|
|
71
|
+
**Yukito Tanabe, Satoru Sugano**
|
|
72
|
+
*Journal of the Physical Society of Japan*, 9, 766-779 (1954)
|
|
73
|
+
**DOI:** 10.1143/JPSJ.9.766
|
|
74
|
+
https://journals.jps.jp/doi/10.1143/JPSJ.9.766
|
|
75
|
+
|
|
76
|
+
3. On the Absorption Spectra of Complex Ions, III The Calculation of the Crystalline Field Strength
|
|
77
|
+
|
|
78
|
+
**Yukito Tanabe, Satoru Sugano**
|
|
79
|
+
*Journal of the Physical Society of Japan*, 11, 864-877 (1956)
|
|
80
|
+
**DOI:** 10.1143/JPSJ.11.864
|
|
81
|
+
https://journals.jps.jp/doi/10.1143/JPSJ.11.864
|
|
82
|
+
|
|
83
|
+
It provides:
|
|
84
|
+
|
|
85
|
+
- Tanabe-Sugano- & Energy-Correlation-Diagrams plotted via `matplotlib`
|
|
86
|
+
- Tanabe-Sugano- & Energy-Correlation-Diagrams exported as `txt`-file
|
|
87
|
+
- Atomic-Termsymbols and their **Eigen-Energies** for a given 10Dq and oxidation state as exported table via `prettytable`
|
|
88
|
+
- Set-up individually **C/B**-ratios
|
|
89
|
+
- Working with Slater-Condon-Parameters **F<sup>2</sup>, F<sup>4</sup>** instead of Racah-Parameters **B, C**
|
|
90
|
+
- Export of the **Tanabe-Sugano-Diagram** as a `html`-file via `plotly` for interactive use
|
|
91
|
+
|
|
92
|
+
The **TanabeSugano**-application can be installed and run:
|
|
93
|
+
|
|
94
|
+
```console
|
|
95
|
+
# via PyPi
|
|
96
|
+
pip install TanabeSugano
|
|
97
|
+
|
|
98
|
+
# via pip+git
|
|
99
|
+
pip git+https://github.com/Anselmoo/TanabeSugano.git
|
|
100
|
+
|
|
101
|
+
# locally
|
|
102
|
+
python setup.py install
|
|
103
|
+
tanabesugano
|
|
104
|
+
|
|
105
|
+
# for plotly-export
|
|
106
|
+
pip install TanabeSugano[plotly]
|
|
107
|
+
```
|
|
108
|
+
|
|
109
|
+
|
|
110
|
+
The options for the **TanabeSugano**-application are:
|
|
111
|
+
|
|
112
|
+
```console
|
|
113
|
+
tanabesugano --help
|
|
114
|
+
|
|
115
|
+
usage: __main__.py [-h] [-d D] [-Dq DQ] [-cut CUT] [-B B B] [-C C C] [-n N]
|
|
116
|
+
[-ndisp] [-ntxt] [-slater]
|
|
117
|
+
|
|
118
|
+
optional arguments:
|
|
119
|
+
-h, --help show this help message and exit
|
|
120
|
+
-d D Number of unpaired electrons (default d5)
|
|
121
|
+
-Dq DQ 10Dq crystal field splitting (default 10Dq = 8065 cm-)
|
|
122
|
+
-cut CUT 10Dq crystal field splitting (default 10Dq = 8065 cm-)
|
|
123
|
+
-B B B Racah Parameter B and the corresponding reduction (default B = 860 cm- * 1.)
|
|
124
|
+
-C C C Racah Parameter C and the corresponding reduction (default C = 4.477*860 cm- * 1.)
|
|
125
|
+
-n N Number of roots (default nroots = 500)
|
|
126
|
+
-ndisp Plot TS-diagram (default = on)
|
|
127
|
+
-ntxt Save TS-diagram and dd energies (default = on)
|
|
128
|
+
-slater Using Slater-Condon F2,F4 parameter instead Racah-Parameter B,C (default = off)
|
|
129
|
+
-v, --version Print version number and exit
|
|
130
|
+
-html Save TS-diagram and dd energies (default = on)
|
|
131
|
+
```
|
|
132
|
+
|
|
133
|
+
**Reference-Example** for d<sup>6</sup> for *B = 860 cm<sup>-</sup>* and *C = 3850 cm<sup>-</sup>* as regular `matplotlib`-plot:
|
|
134
|
+

|
|
135
|
+
|
|
136
|
+
|
|
137
|
+
**Reference-Example** for d<sup>6</sup> for *F<sup>2</sup> = 1065 cm<sup>-</sup>* and *F<sup>4</sup> = 5120 cm<sup>-</sup>* as interactive `plotly`-plot:
|
|
138
|
+
|
|
139
|
+
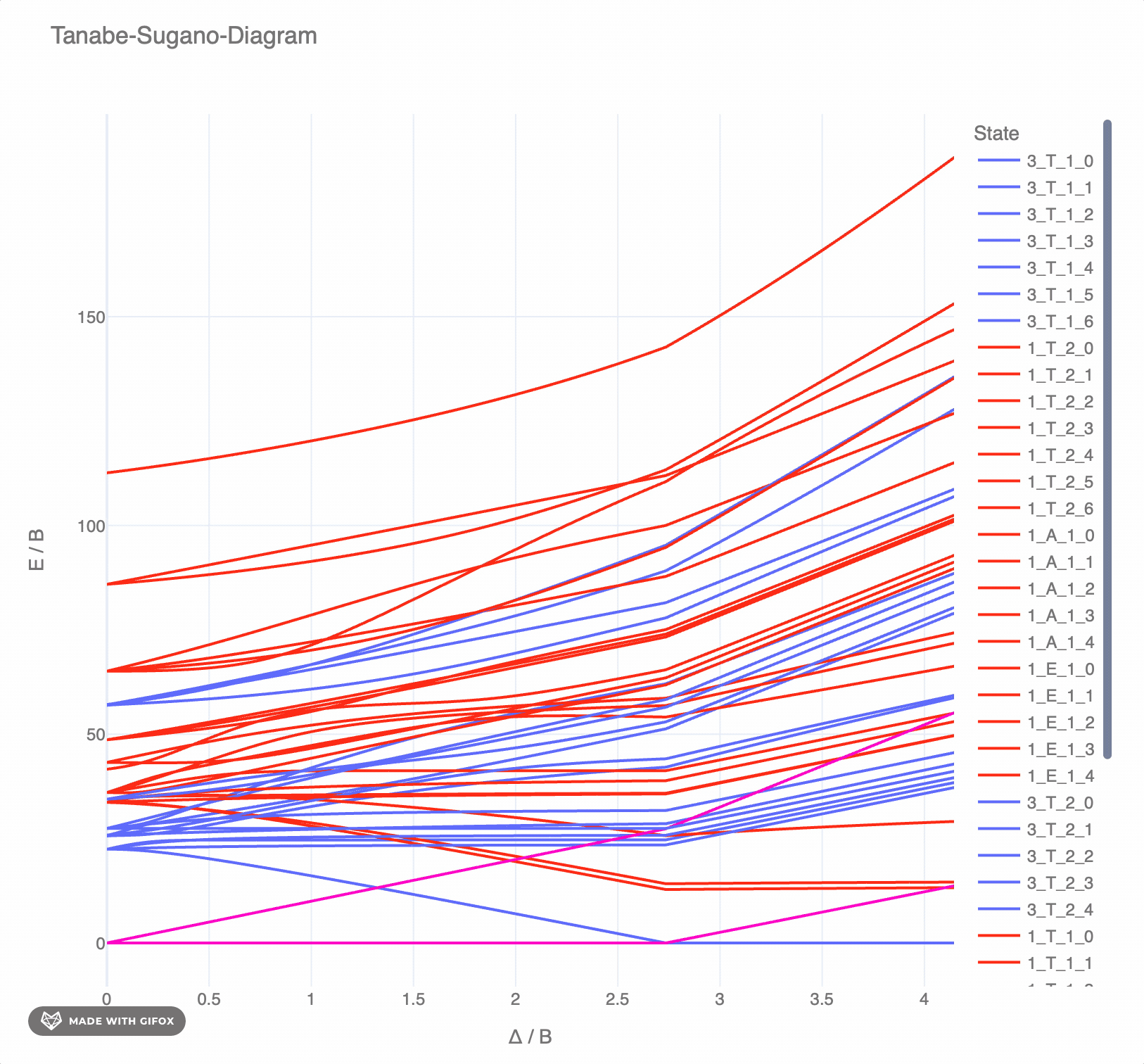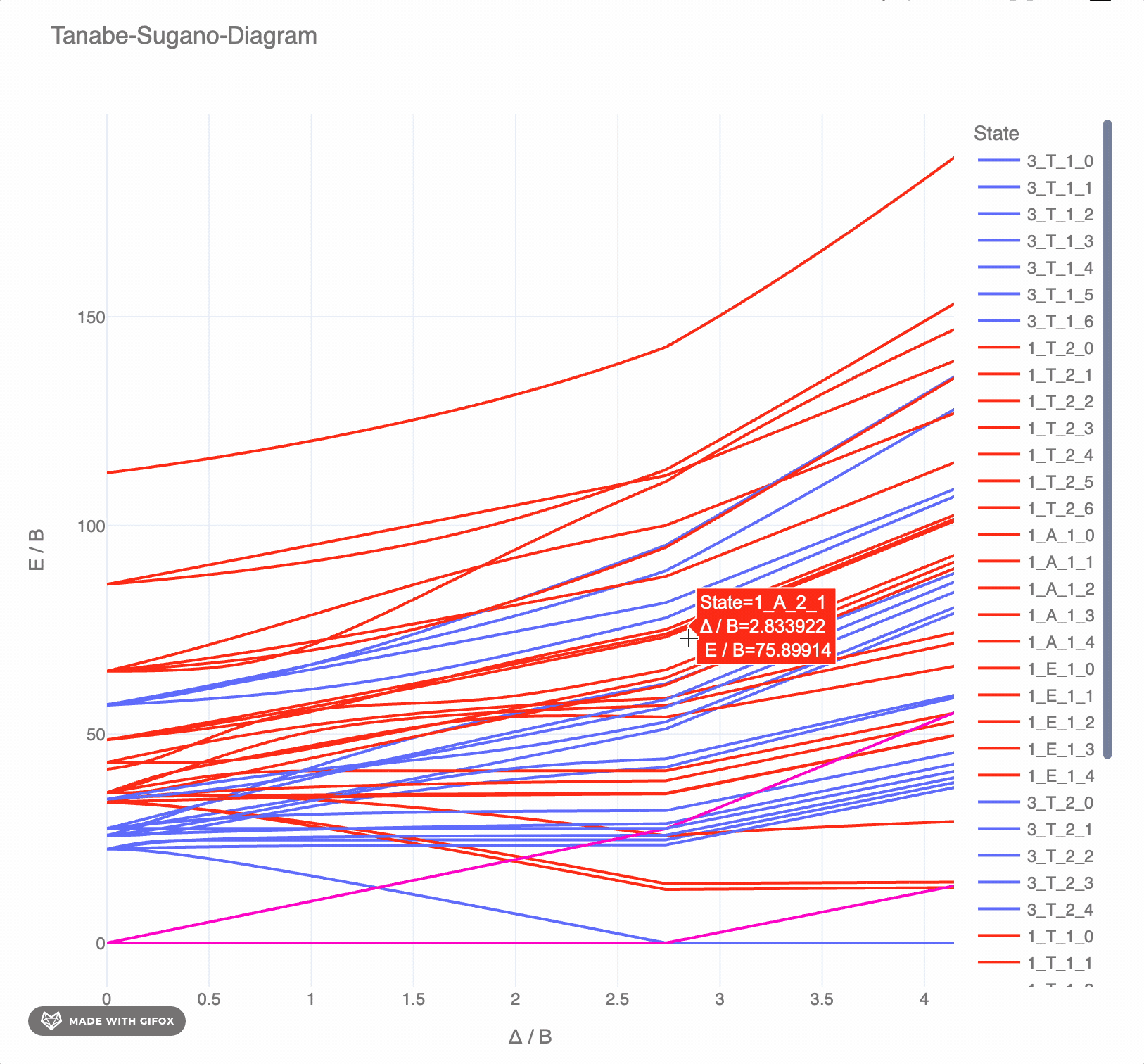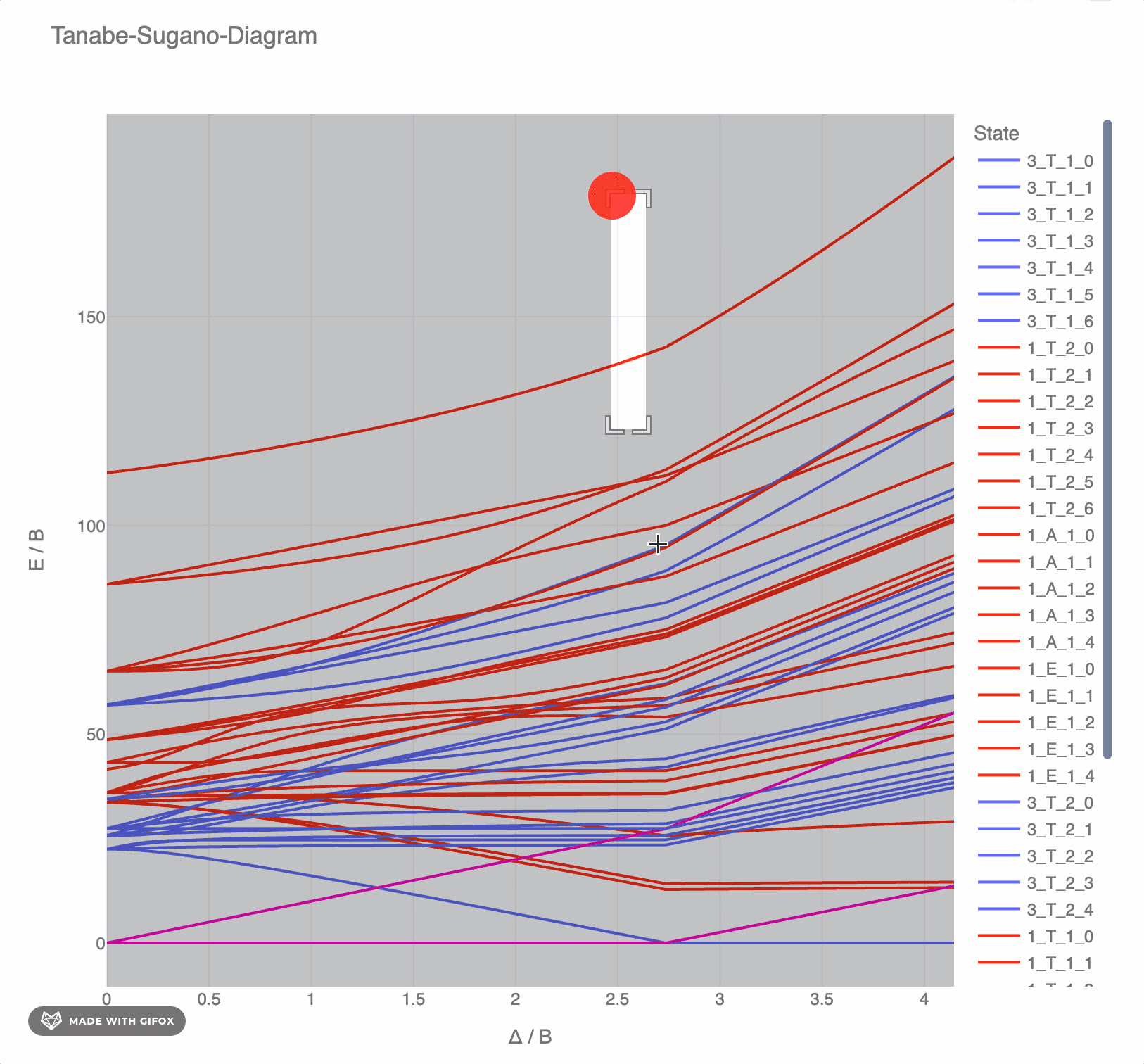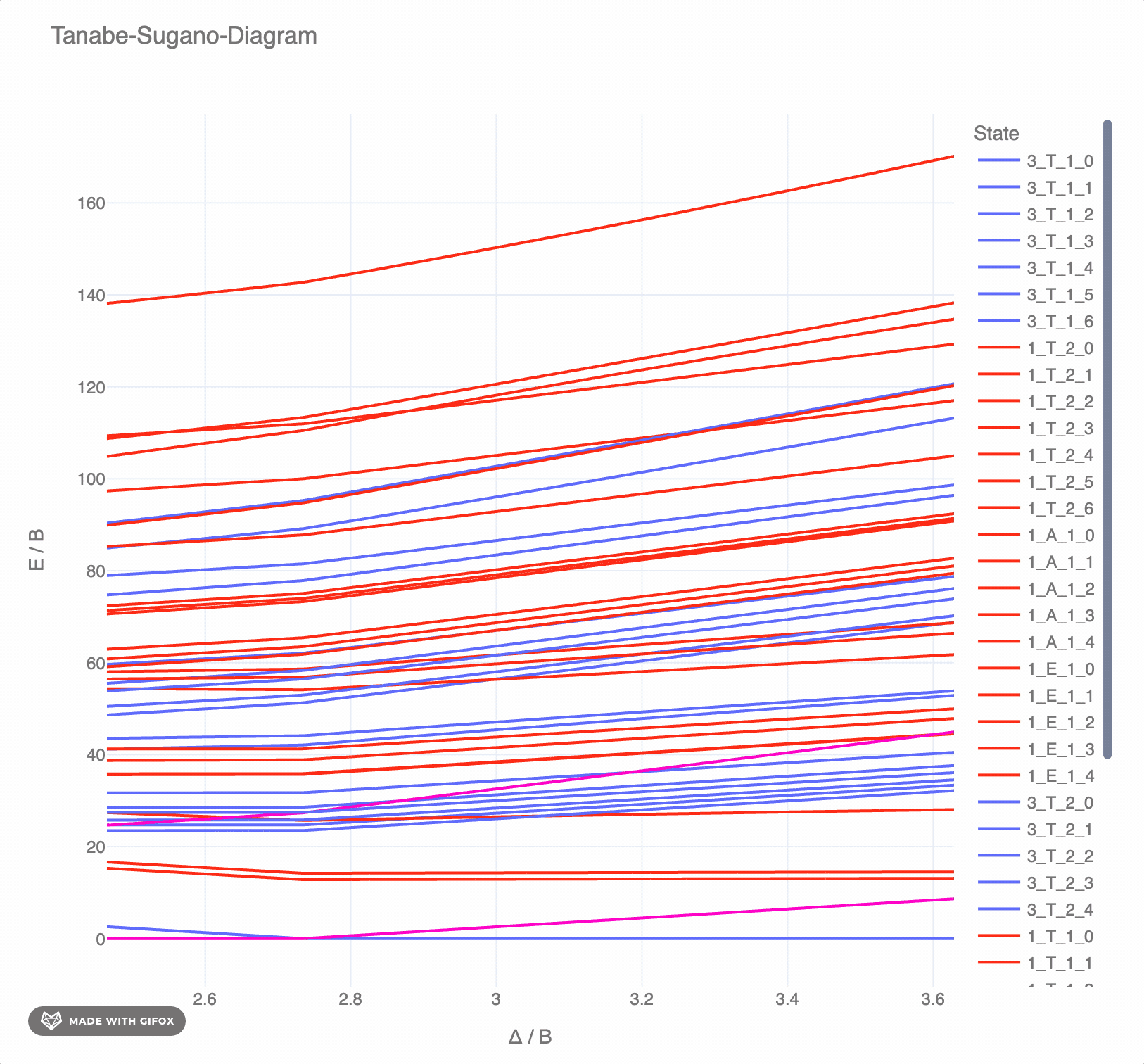
|
|
140
|
+
|
|
141
|
+
## View the TS-diagrams on GitHub Pages
|
|
142
|
+
|
|
143
|
+
All Tanabe-Sugano diagrams (d<sup>2</sup>-d<sup>8</sup>) are available on our interactive GitHub Pages site:
|
|
144
|
+
|
|
145
|
+
**[🔗 View Interactive Diagrams](https://anselmoo.github.io/TanabeSugano/)**
|
|
146
|
+
|
|
147
|
+
## Test the TS-diagrams interactively in Google Colab
|
|
148
|
+
|
|
149
|
+
[](https://colab.research.google.com/github/Anselmoo/TanabeSugano/blob/master/Tanabe_Sugano.ipynb)
|
|
150
|
+
|
|
@@ -0,0 +1,103 @@
|
|
|
1
|
+
[](https://github.com/Anselmoo/TanabeSugano/actions/workflows/python-package.yml)
|
|
2
|
+
[](https://python-poetry.org/)
|
|
3
|
+
[](https://www.codefactor.io/repository/github/anselmoo/tanabesugano)
|
|
4
|
+
[](https://zenodo.org/badge/latestdoi/206847682)
|
|
5
|
+
[](https://github.com/Anselmoo/TanabeSugano/blob/master/LICENSE)
|
|
6
|
+
[](https://github.com/Anselmoo/TanabeSugano/releases)
|
|
7
|
+
[](https://pypi.org/project/TanabeSugano/)
|
|
8
|
+
[](https://pypi.org/project/TanabeSugano/)
|
|
9
|
+
[](https://pepy.tech/project/tanabesugano)
|
|
10
|
+
[](https://colab.research.google.com/github/Anselmoo/TanabeSugano/blob/master/Tanabe_Sugano.ipynb)
|
|
11
|
+
|
|
12
|
+
|
|
13
|
+
# TanabeSugano
|
|
14
|
+
A python-based Eigensolver for Tanabe-Sugano- & Energy-Correlation-Diagrams based on the original three proposed studies of *Yukito Tanabe and Satoru Sugano* for d<sup>2</sup>-d<sup>8</sup> transition metal ions:
|
|
15
|
+
|
|
16
|
+
1. On the Absorption Spectra of Complex Ions. I
|
|
17
|
+
|
|
18
|
+
**Yukito Tanabe, Satoru Sugano**
|
|
19
|
+
*Journal of the Physical Society of Japan*, 9, 753-766 (1954)
|
|
20
|
+
**DOI:** 10.1143/JPSJ.9.753
|
|
21
|
+
https://journals.jps.jp/doi/10.1143/JPSJ.9.753
|
|
22
|
+
|
|
23
|
+
2. On the Absorption Spectra of Complex Ions II
|
|
24
|
+
|
|
25
|
+
**Yukito Tanabe, Satoru Sugano**
|
|
26
|
+
*Journal of the Physical Society of Japan*, 9, 766-779 (1954)
|
|
27
|
+
**DOI:** 10.1143/JPSJ.9.766
|
|
28
|
+
https://journals.jps.jp/doi/10.1143/JPSJ.9.766
|
|
29
|
+
|
|
30
|
+
3. On the Absorption Spectra of Complex Ions, III The Calculation of the Crystalline Field Strength
|
|
31
|
+
|
|
32
|
+
**Yukito Tanabe, Satoru Sugano**
|
|
33
|
+
*Journal of the Physical Society of Japan*, 11, 864-877 (1956)
|
|
34
|
+
**DOI:** 10.1143/JPSJ.11.864
|
|
35
|
+
https://journals.jps.jp/doi/10.1143/JPSJ.11.864
|
|
36
|
+
|
|
37
|
+
It provides:
|
|
38
|
+
|
|
39
|
+
- Tanabe-Sugano- & Energy-Correlation-Diagrams plotted via `matplotlib`
|
|
40
|
+
- Tanabe-Sugano- & Energy-Correlation-Diagrams exported as `txt`-file
|
|
41
|
+
- Atomic-Termsymbols and their **Eigen-Energies** for a given 10Dq and oxidation state as exported table via `prettytable`
|
|
42
|
+
- Set-up individually **C/B**-ratios
|
|
43
|
+
- Working with Slater-Condon-Parameters **F<sup>2</sup>, F<sup>4</sup>** instead of Racah-Parameters **B, C**
|
|
44
|
+
- Export of the **Tanabe-Sugano-Diagram** as a `html`-file via `plotly` for interactive use
|
|
45
|
+
|
|
46
|
+
The **TanabeSugano**-application can be installed and run:
|
|
47
|
+
|
|
48
|
+
```console
|
|
49
|
+
# via PyPi
|
|
50
|
+
pip install TanabeSugano
|
|
51
|
+
|
|
52
|
+
# via pip+git
|
|
53
|
+
pip git+https://github.com/Anselmoo/TanabeSugano.git
|
|
54
|
+
|
|
55
|
+
# locally
|
|
56
|
+
python setup.py install
|
|
57
|
+
tanabesugano
|
|
58
|
+
|
|
59
|
+
# for plotly-export
|
|
60
|
+
pip install TanabeSugano[plotly]
|
|
61
|
+
```
|
|
62
|
+
|
|
63
|
+
|
|
64
|
+
The options for the **TanabeSugano**-application are:
|
|
65
|
+
|
|
66
|
+
```console
|
|
67
|
+
tanabesugano --help
|
|
68
|
+
|
|
69
|
+
usage: __main__.py [-h] [-d D] [-Dq DQ] [-cut CUT] [-B B B] [-C C C] [-n N]
|
|
70
|
+
[-ndisp] [-ntxt] [-slater]
|
|
71
|
+
|
|
72
|
+
optional arguments:
|
|
73
|
+
-h, --help show this help message and exit
|
|
74
|
+
-d D Number of unpaired electrons (default d5)
|
|
75
|
+
-Dq DQ 10Dq crystal field splitting (default 10Dq = 8065 cm-)
|
|
76
|
+
-cut CUT 10Dq crystal field splitting (default 10Dq = 8065 cm-)
|
|
77
|
+
-B B B Racah Parameter B and the corresponding reduction (default B = 860 cm- * 1.)
|
|
78
|
+
-C C C Racah Parameter C and the corresponding reduction (default C = 4.477*860 cm- * 1.)
|
|
79
|
+
-n N Number of roots (default nroots = 500)
|
|
80
|
+
-ndisp Plot TS-diagram (default = on)
|
|
81
|
+
-ntxt Save TS-diagram and dd energies (default = on)
|
|
82
|
+
-slater Using Slater-Condon F2,F4 parameter instead Racah-Parameter B,C (default = off)
|
|
83
|
+
-v, --version Print version number and exit
|
|
84
|
+
-html Save TS-diagram and dd energies (default = on)
|
|
85
|
+
```
|
|
86
|
+
|
|
87
|
+
**Reference-Example** for d<sup>6</sup> for *B = 860 cm<sup>-</sup>* and *C = 3850 cm<sup>-</sup>* as regular `matplotlib`-plot:
|
|
88
|
+

|
|
89
|
+
|
|
90
|
+
|
|
91
|
+
**Reference-Example** for d<sup>6</sup> for *F<sup>2</sup> = 1065 cm<sup>-</sup>* and *F<sup>4</sup> = 5120 cm<sup>-</sup>* as interactive `plotly`-plot:
|
|
92
|
+
|
|
93
|
+

|
|
94
|
+
|
|
95
|
+
## View the TS-diagrams on GitHub Pages
|
|
96
|
+
|
|
97
|
+
All Tanabe-Sugano diagrams (d<sup>2</sup>-d<sup>8</sup>) are available on our interactive GitHub Pages site:
|
|
98
|
+
|
|
99
|
+
**[🔗 View Interactive Diagrams](https://anselmoo.github.io/TanabeSugano/)**
|
|
100
|
+
|
|
101
|
+
## Test the TS-diagrams interactively in Google Colab
|
|
102
|
+
|
|
103
|
+
[](https://colab.research.google.com/github/Anselmoo/TanabeSugano/blob/master/Tanabe_Sugano.ipynb)
|
|
@@ -0,0 +1,97 @@
|
|
|
1
|
+
[tool.poetry]
|
|
2
|
+
name = "tanabesugano"
|
|
3
|
+
version = "1.5.0"
|
|
4
|
+
description = "A python-solver for Tanabe-Sugano and Energy-Correlation diagrams"
|
|
5
|
+
authors = ["Anselm Hahn <anselm.hahn@gmail.com>"]
|
|
6
|
+
maintainers = ["Anselm Hahn <anselm.hahn@gmail.com>"]
|
|
7
|
+
readme = "README.md"
|
|
8
|
+
license = "MIT"
|
|
9
|
+
keywords = [
|
|
10
|
+
"terminal",
|
|
11
|
+
"data-visualization",
|
|
12
|
+
"tanabe-sugano",
|
|
13
|
+
"energy-correlation",
|
|
14
|
+
"complex-ions",
|
|
15
|
+
]
|
|
16
|
+
repository = "https://github.com/Anselmoo/TanabeSugano"
|
|
17
|
+
homepage = "https://pypi.org/project/TanabeSugano"
|
|
18
|
+
classifiers = [
|
|
19
|
+
"Development Status :: 5 - Production/Stable",
|
|
20
|
+
"Environment :: Console",
|
|
21
|
+
"Intended Audience :: Education",
|
|
22
|
+
"Intended Audience :: Science/Research",
|
|
23
|
+
"License :: OSI Approved :: MIT License",
|
|
24
|
+
"Operating System :: MacOS",
|
|
25
|
+
"Operating System :: Unix",
|
|
26
|
+
"Operating System :: Microsoft :: Windows",
|
|
27
|
+
"Programming Language :: Python :: 3",
|
|
28
|
+
"Programming Language :: Python :: 3.8",
|
|
29
|
+
"Programming Language :: Python :: 3.9",
|
|
30
|
+
"Programming Language :: Python :: 3.10",
|
|
31
|
+
"Programming Language :: Python :: 3.11",
|
|
32
|
+
"Programming Language :: Python :: 3.12",
|
|
33
|
+
"Topic :: System :: Shells",
|
|
34
|
+
"Topic :: Scientific/Engineering :: Visualization",
|
|
35
|
+
"Topic :: Scientific/Engineering :: Chemistry",
|
|
36
|
+
"Topic :: Scientific/Engineering :: Physics",
|
|
37
|
+
"Topic :: Utilities",
|
|
38
|
+
]
|
|
39
|
+
include = ["LICENSE"]
|
|
40
|
+
exclude = ["test/**/*.py", "example/**", "ts-diagrams/**"]
|
|
41
|
+
|
|
42
|
+
[tool.poetry.dependencies]
|
|
43
|
+
python = ">=3.9,<4.0"
|
|
44
|
+
pandas = ">=1.2.4,<4.0.0"
|
|
45
|
+
numpy = [
|
|
46
|
+
{ version = "^1.24.4", python = ">=3.8,<3.9" },
|
|
47
|
+
{ version = "^1.26.4", python = ">=3.9,<3.10" },
|
|
48
|
+
{ version = "^2.0.2", python = ">=3.10,<3.13" },
|
|
49
|
+
]
|
|
50
|
+
matplotlib = "^3.4.2"
|
|
51
|
+
prettytable = ">=2.1,<4.0"
|
|
52
|
+
plotly = { version = ">=5.13.1,<7.0.0", optional = true }
|
|
53
|
+
update = "^0.0.1"
|
|
54
|
+
|
|
55
|
+
[tool.poetry.group.dev.dependencies]
|
|
56
|
+
pytest = ">=7.2,<9.0"
|
|
57
|
+
pytest-clarity = "^1.0.1"
|
|
58
|
+
pytest-cov = ">=4,<6"
|
|
59
|
+
flake8 = ">=6,<8"
|
|
60
|
+
black = ">=22.12,<25.0"
|
|
61
|
+
isort = "^5.11.4"
|
|
62
|
+
pytest-console-scripts = "^1.3.1"
|
|
63
|
+
ruff = "^0.7.0"
|
|
64
|
+
|
|
65
|
+
[build-system]
|
|
66
|
+
requires = ["poetry-core>=1.1.0"]
|
|
67
|
+
build-backend = "poetry.core.masonry.api"
|
|
68
|
+
|
|
69
|
+
[tool.poetry.scripts]
|
|
70
|
+
tanabesugano = "tanabesugano.cmd:cmd_line"
|
|
71
|
+
|
|
72
|
+
[tool.poetry.extras]
|
|
73
|
+
plotly = ["plotly"]
|
|
74
|
+
|
|
75
|
+
[tool.ruff]
|
|
76
|
+
lint.select = ["ALL"]
|
|
77
|
+
lint.ignore = ["UP006", "UP007", "N801", "N802", "N803", "N806"]
|
|
78
|
+
target-version = "py38"
|
|
79
|
+
src = ["tanabesugano"]
|
|
80
|
+
|
|
81
|
+
[tool.ruff.lint.per-file-ignores]
|
|
82
|
+
"tanabesugano/test/*" = ["PT006", "ANN001", "ANN201", "D103", "PLR2004", "S101"]
|
|
83
|
+
|
|
84
|
+
[tool.ruff.lint.isort]
|
|
85
|
+
known-first-party = ["umf"]
|
|
86
|
+
force-single-line = true
|
|
87
|
+
lines-between-types = 1
|
|
88
|
+
lines-after-imports = 2
|
|
89
|
+
known-third-party = ["poetry.core"]
|
|
90
|
+
required-imports = ["from __future__ import annotations"]
|
|
91
|
+
|
|
92
|
+
[tool.pytest.ini_options]
|
|
93
|
+
markers = """
|
|
94
|
+
xfail: mark test as expecting to fail
|
|
95
|
+
skip: mark test as skipped
|
|
96
|
+
"""
|
|
97
|
+
addopts = "-v --cov=tanabesugano"
|
|
@@ -0,0 +1,159 @@
|
|
|
1
|
+
from __future__ import annotations
|
|
2
|
+
|
|
3
|
+
from typing import List
|
|
4
|
+
|
|
5
|
+
import numpy as np
|
|
6
|
+
|
|
7
|
+
from tanabesugano import matrices
|
|
8
|
+
from tanabesugano import tools
|
|
9
|
+
|
|
10
|
+
|
|
11
|
+
class Batch:
|
|
12
|
+
def __init__(
|
|
13
|
+
self,
|
|
14
|
+
Dq: List[float] = None,
|
|
15
|
+
B: List[float] = None,
|
|
16
|
+
C: List[float] = None,
|
|
17
|
+
d_count: int = 5,
|
|
18
|
+
slater: bool = False,
|
|
19
|
+
):
|
|
20
|
+
if Dq is None:
|
|
21
|
+
Dq = [4000.0, 4500.0, 10]
|
|
22
|
+
if B is None:
|
|
23
|
+
B = [400.0, 4500.0, 10]
|
|
24
|
+
if C is None:
|
|
25
|
+
C = [3600.0, 4000, 10]
|
|
26
|
+
if len(Dq) != 3:
|
|
27
|
+
raise KeyError(
|
|
28
|
+
"The range of `Dq` is based on the three values: start, stop, steps!",
|
|
29
|
+
)
|
|
30
|
+
self.Dq = np.linspace(Dq[0], Dq[1], int(Dq[2])) # Oh-crystalfield-splitting
|
|
31
|
+
if len(B) != 3:
|
|
32
|
+
raise KeyError(
|
|
33
|
+
"The range of `B` is based on the three values: start, stop, steps!",
|
|
34
|
+
)
|
|
35
|
+
self.B = np.linspace(
|
|
36
|
+
B[0],
|
|
37
|
+
B[1],
|
|
38
|
+
int(B[2]),
|
|
39
|
+
) # Racah-Parameter B in wavenumbers
|
|
40
|
+
if len(C) != 3:
|
|
41
|
+
raise KeyError(
|
|
42
|
+
"The range of `C` is based on the three values: start, stop, steps!",
|
|
43
|
+
)
|
|
44
|
+
self.C = np.linspace(
|
|
45
|
+
C[0],
|
|
46
|
+
C[1],
|
|
47
|
+
int(C[2]),
|
|
48
|
+
) # Racah-Parameter C in wavenumbers
|
|
49
|
+
|
|
50
|
+
if slater:
|
|
51
|
+
# Transformin Racah to Slater-Condon
|
|
52
|
+
self.B, self.C = tools.racah(B, C)
|
|
53
|
+
|
|
54
|
+
# self.delta_B = self.e_range / self.B
|
|
55
|
+
|
|
56
|
+
self.d_count = d_count
|
|
57
|
+
if self.d_count in {4, 5, 6}:
|
|
58
|
+
self._size = 42
|
|
59
|
+
if self.d_count in {3, 7}:
|
|
60
|
+
self._size = 19
|
|
61
|
+
if self.d_count in {2, 8}:
|
|
62
|
+
self._size = 10
|
|
63
|
+
self.result: List[dict] = []
|
|
64
|
+
|
|
65
|
+
def calculation(self) -> None:
|
|
66
|
+
"""Is filling the self.result with the iTS states of over-iterated energy range."""
|
|
67
|
+
for _Dq in self.Dq:
|
|
68
|
+
for _B in self.B:
|
|
69
|
+
for _C in self.C:
|
|
70
|
+
if self.d_count == 2: # d2
|
|
71
|
+
states = matrices.d2(Dq=_Dq, B=_B, C=_C).solver()
|
|
72
|
+
self.result.append(
|
|
73
|
+
{
|
|
74
|
+
"d_count": self.d_count,
|
|
75
|
+
"Dq": _Dq,
|
|
76
|
+
"B": _B,
|
|
77
|
+
"C": _C,
|
|
78
|
+
"states": states,
|
|
79
|
+
},
|
|
80
|
+
)
|
|
81
|
+
|
|
82
|
+
elif self.d_count == 3: # d3
|
|
83
|
+
states = matrices.d3(Dq=_Dq, B=_B, C=_C).solver()
|
|
84
|
+
self.result.append(
|
|
85
|
+
{
|
|
86
|
+
"d_count": self.d_count,
|
|
87
|
+
"Dq": _Dq,
|
|
88
|
+
"B": _B,
|
|
89
|
+
"C": _C,
|
|
90
|
+
"states": states,
|
|
91
|
+
},
|
|
92
|
+
)
|
|
93
|
+
elif self.d_count == 4: # d4
|
|
94
|
+
states = matrices.d4(Dq=_Dq, B=_B, C=_C).solver()
|
|
95
|
+
self.result.append(
|
|
96
|
+
{
|
|
97
|
+
"d_count": self.d_count,
|
|
98
|
+
"Dq": _Dq,
|
|
99
|
+
"B": _B,
|
|
100
|
+
"C": _C,
|
|
101
|
+
"states": states,
|
|
102
|
+
},
|
|
103
|
+
)
|
|
104
|
+
elif self.d_count == 5: # d5
|
|
105
|
+
states = matrices.d5(Dq=_Dq, B=_B, C=_C).solver()
|
|
106
|
+
self.result.append(
|
|
107
|
+
{
|
|
108
|
+
"d_count": self.d_count,
|
|
109
|
+
"Dq": _Dq,
|
|
110
|
+
"B": _B,
|
|
111
|
+
"C": _C,
|
|
112
|
+
"states": states,
|
|
113
|
+
},
|
|
114
|
+
)
|
|
115
|
+
elif self.d_count == 6: # d6
|
|
116
|
+
states = matrices.d6(Dq=_Dq, B=_B, C=_C).solver()
|
|
117
|
+
self.result.append(
|
|
118
|
+
{
|
|
119
|
+
"d_count": self.d_count,
|
|
120
|
+
"Dq": _Dq,
|
|
121
|
+
"B": _B,
|
|
122
|
+
"C": _C,
|
|
123
|
+
"states": states,
|
|
124
|
+
},
|
|
125
|
+
)
|
|
126
|
+
elif self.d_count == 7: # d7
|
|
127
|
+
states = matrices.d7(Dq=_Dq, B=_B, C=_C).solver()
|
|
128
|
+
self.result.append(
|
|
129
|
+
{
|
|
130
|
+
"d_count": self.d_count,
|
|
131
|
+
"Dq": _Dq,
|
|
132
|
+
"B": _B,
|
|
133
|
+
"C": _C,
|
|
134
|
+
"states": states,
|
|
135
|
+
},
|
|
136
|
+
)
|
|
137
|
+
elif self.d_count == 8: # d8
|
|
138
|
+
states = matrices.d8(Dq=_Dq, B=_B, C=_C).solver()
|
|
139
|
+
self.result.append(
|
|
140
|
+
{
|
|
141
|
+
"d_count": self.d_count,
|
|
142
|
+
"Dq": _Dq,
|
|
143
|
+
"B": _B,
|
|
144
|
+
"C": _C,
|
|
145
|
+
"states": states,
|
|
146
|
+
},
|
|
147
|
+
)
|
|
148
|
+
else:
|
|
149
|
+
raise ValueError("not a correct value!")
|
|
150
|
+
|
|
151
|
+
@property
|
|
152
|
+
def return_result(self) -> List[dict]:
|
|
153
|
+
return self.result
|
|
154
|
+
|
|
155
|
+
|
|
156
|
+
if __name__ == "__main__":
|
|
157
|
+
res = Batch()
|
|
158
|
+
res.calculation()
|
|
159
|
+
print(res.return_result)
|