pydartdiags 0.0.4__tar.gz → 0.0.42__tar.gz
This diff represents the content of publicly available package versions that have been released to one of the supported registries. The information contained in this diff is provided for informational purposes only and reflects changes between package versions as they appear in their respective public registries.
Potentially problematic release.
This version of pydartdiags might be problematic. Click here for more details.
- {pydartdiags-0.0.4 → pydartdiags-0.0.42}/PKG-INFO +29 -18
- {pydartdiags-0.0.4 → pydartdiags-0.0.42}/README.md +19 -11
- {pydartdiags-0.0.4 → pydartdiags-0.0.42}/pyproject.toml +5 -6
- pydartdiags-0.0.42/setup.cfg +4 -0
- pydartdiags-0.0.42/setup.py +26 -0
- {pydartdiags-0.0.4 → pydartdiags-0.0.42}/src/pydartdiags/obs_sequence/obs_sequence.py +270 -14
- {pydartdiags-0.0.4 → pydartdiags-0.0.42}/src/pydartdiags/plots/plots.py +3 -1
- pydartdiags-0.0.42/src/pydartdiags.egg-info/PKG-INFO +404 -0
- pydartdiags-0.0.42/src/pydartdiags.egg-info/SOURCES.txt +16 -0
- pydartdiags-0.0.42/src/pydartdiags.egg-info/dependency_links.txt +1 -0
- pydartdiags-0.0.42/src/pydartdiags.egg-info/requires.txt +4 -0
- pydartdiags-0.0.42/src/pydartdiags.egg-info/top_level.txt +1 -0
- pydartdiags-0.0.42/tests/test_obs_sequence.py +87 -0
- pydartdiags-0.0.42/tests/test_plots.py +52 -0
- pydartdiags-0.0.4/.gitignore +0 -4
- pydartdiags-0.0.4/docs/.nojekyll +0 -0
- pydartdiags-0.0.4/docs/Makefile +0 -20
- pydartdiags-0.0.4/docs/build/doctrees/environment.pickle +0 -0
- pydartdiags-0.0.4/docs/build/doctrees/index.doctree +0 -0
- pydartdiags-0.0.4/docs/build/doctrees/obs_sequence.doctree +0 -0
- pydartdiags-0.0.4/docs/build/doctrees/plots.doctree +0 -0
- pydartdiags-0.0.4/docs/build/doctrees/quickstart.doctree +0 -0
- pydartdiags-0.0.4/docs/build/html/.buildinfo +0 -4
- pydartdiags-0.0.4/docs/build/html/_images/bias.png +0 -0
- pydartdiags-0.0.4/docs/build/html/_images/rankhist.png +0 -0
- pydartdiags-0.0.4/docs/build/html/_images/rmse.png +0 -0
- pydartdiags-0.0.4/docs/build/html/_sources/index.rst.txt +0 -40
- pydartdiags-0.0.4/docs/build/html/_sources/obs_sequence.rst.txt +0 -8
- pydartdiags-0.0.4/docs/build/html/_sources/plots.rst.txt +0 -6
- pydartdiags-0.0.4/docs/build/html/_sources/quickstart.rst.txt +0 -380
- pydartdiags-0.0.4/docs/build/html/_static/alabaster.css +0 -714
- pydartdiags-0.0.4/docs/build/html/_static/basic.css +0 -925
- pydartdiags-0.0.4/docs/build/html/_static/check-solid.svg +0 -4
- pydartdiags-0.0.4/docs/build/html/_static/clipboard.min.js +0 -7
- pydartdiags-0.0.4/docs/build/html/_static/copy-button.svg +0 -5
- pydartdiags-0.0.4/docs/build/html/_static/copybutton.css +0 -94
- pydartdiags-0.0.4/docs/build/html/_static/copybutton.js +0 -248
- pydartdiags-0.0.4/docs/build/html/_static/copybutton_funcs.js +0 -73
- pydartdiags-0.0.4/docs/build/html/_static/custom.css +0 -1
- pydartdiags-0.0.4/docs/build/html/_static/doctools.js +0 -156
- pydartdiags-0.0.4/docs/build/html/_static/documentation_options.js +0 -13
- pydartdiags-0.0.4/docs/build/html/_static/file.png +0 -0
- pydartdiags-0.0.4/docs/build/html/_static/language_data.js +0 -199
- pydartdiags-0.0.4/docs/build/html/_static/minus.png +0 -0
- pydartdiags-0.0.4/docs/build/html/_static/plus.png +0 -0
- pydartdiags-0.0.4/docs/build/html/_static/pygments.css +0 -84
- pydartdiags-0.0.4/docs/build/html/_static/searchtools.js +0 -620
- pydartdiags-0.0.4/docs/build/html/_static/sphinx_highlight.js +0 -154
- pydartdiags-0.0.4/docs/build/html/genindex.html +0 -297
- pydartdiags-0.0.4/docs/build/html/index.html +0 -131
- pydartdiags-0.0.4/docs/build/html/objects.inv +0 -0
- pydartdiags-0.0.4/docs/build/html/obs_sequence.html +0 -479
- pydartdiags-0.0.4/docs/build/html/plots.html +0 -240
- pydartdiags-0.0.4/docs/build/html/py-modindex.html +0 -137
- pydartdiags-0.0.4/docs/build/html/quickstart.html +0 -471
- pydartdiags-0.0.4/docs/build/html/search.html +0 -127
- pydartdiags-0.0.4/docs/build/html/searchindex.js +0 -1
- pydartdiags-0.0.4/docs/conf.py +0 -60
- pydartdiags-0.0.4/docs/images/bias.png +0 -0
- pydartdiags-0.0.4/docs/images/rankhist.png +0 -0
- pydartdiags-0.0.4/docs/images/rmse.png +0 -0
- pydartdiags-0.0.4/docs/index.html +0 -1
- pydartdiags-0.0.4/docs/index.rst +0 -40
- pydartdiags-0.0.4/docs/make.bat +0 -35
- pydartdiags-0.0.4/docs/obs_sequence.rst +0 -8
- pydartdiags-0.0.4/docs/plots.rst +0 -6
- pydartdiags-0.0.4/docs/quickstart.rst +0 -380
- pydartdiags-0.0.4/src/pydartdiags/obs_sequence/composite_types.yaml +0 -35
- pydartdiags-0.0.4/src/pydartdiags/plots/tests/test_rank_histogram.py +0 -18
- {pydartdiags-0.0.4 → pydartdiags-0.0.42}/LICENSE +0 -0
- {pydartdiags-0.0.4 → pydartdiags-0.0.42}/src/pydartdiags/__init__.py +0 -0
- {pydartdiags-0.0.4 → pydartdiags-0.0.42}/src/pydartdiags/obs_sequence/__init__.py +0 -0
- {pydartdiags-0.0.4 → pydartdiags-0.0.42}/src/pydartdiags/plots/__init__.py +0 -0
|
@@ -1,30 +1,41 @@
|
|
|
1
|
-
Metadata-Version: 2.
|
|
1
|
+
Metadata-Version: 2.1
|
|
2
2
|
Name: pydartdiags
|
|
3
|
-
Version: 0.0.
|
|
3
|
+
Version: 0.0.42
|
|
4
4
|
Summary: Observation Sequence Diagnostics for DART
|
|
5
|
+
Home-page: https://github.com/NCAR/pyDARTdiags.git
|
|
6
|
+
Author: Helen Kershaw
|
|
7
|
+
Author-email: Helen Kershaw <hkershaw@ucar.edu>
|
|
5
8
|
Project-URL: Homepage, https://github.com/NCAR/pyDARTdiags.git
|
|
6
9
|
Project-URL: Issues, https://github.com/NCAR/pyDARTdiags/issues
|
|
7
10
|
Project-URL: Documentation, https://ncar.github.io/pyDARTdiags
|
|
8
|
-
|
|
9
|
-
License-File: LICENSE
|
|
11
|
+
Classifier: Programming Language :: Python :: 3
|
|
10
12
|
Classifier: License :: OSI Approved :: Apache Software License
|
|
11
13
|
Classifier: Operating System :: OS Independent
|
|
12
|
-
Classifier: Programming Language :: Python :: 3
|
|
13
14
|
Requires-Python: >=3.8
|
|
14
|
-
|
|
15
|
+
Description-Content-Type: text/markdown
|
|
16
|
+
License-File: LICENSE
|
|
15
17
|
Requires-Dist: pandas>=2.2.0
|
|
18
|
+
Requires-Dist: numpy>=1.26
|
|
16
19
|
Requires-Dist: plotly>=5.22.0
|
|
17
|
-
|
|
20
|
+
Requires-Dist: pyyaml>=6.0.2
|
|
21
|
+
|
|
22
|
+
[](https://opensource.org/licenses/Apache-2.0)
|
|
23
|
+
[](https://codecov.io/gh/NCAR/pyDARTdiags)
|
|
24
|
+
[](https://pypi.org/project/pydartdiags/)
|
|
25
|
+
|
|
18
26
|
|
|
19
27
|
# pyDARTdiags
|
|
20
28
|
|
|
21
|
-
pyDARTdiags is a
|
|
29
|
+
pyDARTdiags is a Python library for obsevation space diagnostics for the Data Assimilation Research Testbed ([DART](https://github.com/NCAR/DART)).
|
|
22
30
|
|
|
23
31
|
pyDARTdiags is under initial development, so please use caution.
|
|
24
32
|
The MATLAB [observation space diagnostics](https://docs.dart.ucar.edu/en/latest/guide/matlab-observation-space.html) are available through [DART](https://github.com/NCAR/DART).
|
|
25
33
|
|
|
26
34
|
|
|
27
|
-
pyDARTdiags can be installed through pip.
|
|
35
|
+
pyDARTdiags can be installed through pip: https://pypi.org/project/pydartdiags/
|
|
36
|
+
Documenation : https://ncar.github.io/pyDARTdiags/
|
|
37
|
+
|
|
38
|
+
We recommend installing pydartdiags in a virtual enviroment:
|
|
28
39
|
|
|
29
40
|
|
|
30
41
|
```
|
|
@@ -36,14 +47,14 @@ pip install pydartdiags
|
|
|
36
47
|
## Example importing the obs\_sequence and plots modules
|
|
37
48
|
|
|
38
49
|
```python
|
|
39
|
-
from pydartdiags.obs_sequence import obs_sequence as
|
|
50
|
+
from pydartdiags.obs_sequence import obs_sequence as obsq
|
|
40
51
|
from pydartdiags.plots import plots
|
|
41
52
|
```
|
|
42
53
|
|
|
43
54
|
## Examining the dataframe
|
|
44
55
|
|
|
45
56
|
```python
|
|
46
|
-
obs_seq =
|
|
57
|
+
obs_seq = obsq.obs_sequence('obs_seq.final.ascii')
|
|
47
58
|
obs_seq.df.head()
|
|
48
59
|
```
|
|
49
60
|
|
|
@@ -204,7 +215,7 @@ obs_seq.df.head()
|
|
|
204
215
|
Find the numeber of assimilated (used) observations vs. possible observations by type
|
|
205
216
|
|
|
206
217
|
```python
|
|
207
|
-
|
|
218
|
+
obsq.possible_vs_used(obs_seq.df)
|
|
208
219
|
```
|
|
209
220
|
|
|
210
221
|
<table border="1" class="dataframe">
|
|
@@ -361,10 +372,10 @@ obs_seq.possible_vs_used(obs_seq.df)
|
|
|
361
372
|
* plot the rank histogram
|
|
362
373
|
|
|
363
374
|
```python
|
|
364
|
-
df_qc0 =
|
|
375
|
+
df_qc0 = obsq.select_by_dart_qc(obs_seq.df, 0)
|
|
365
376
|
plots.plot_rank_histogram(df_qc0)
|
|
366
377
|
```
|
|
367
|
-

|
|
378
|
+
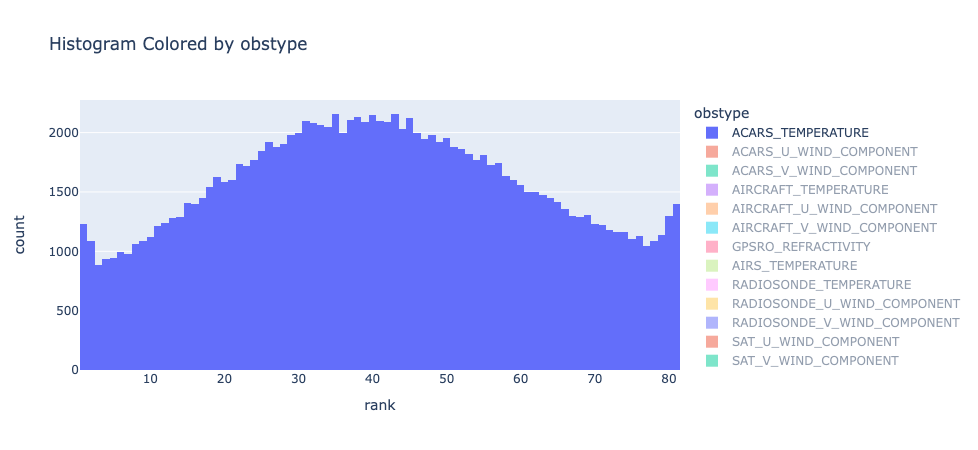
|
|
368
379
|
|
|
369
380
|
|
|
370
381
|
### plot profile of RMSE and Bias
|
|
@@ -377,17 +388,17 @@ plots.plot_rank_histogram(df_qc0)
|
|
|
377
388
|
hPalevels = [0.0, 100.0, 150.0, 200.0, 250.0, 300.0, 400.0, 500.0, 700, 850, 925, 1000]# float("inf")] # Pa?
|
|
378
389
|
plevels = [i * 100 for i in hPalevels]
|
|
379
390
|
|
|
380
|
-
df_qc0 =
|
|
391
|
+
df_qc0 = obsq.select_by_dart_qc(obs_seq.df, 0) # only qc 0
|
|
381
392
|
df_profile, figrmse, figbias = plots.plot_profile(df_qc0, plevels)
|
|
382
393
|
```
|
|
383
394
|
|
|
384
|
-

|
|
395
|
+
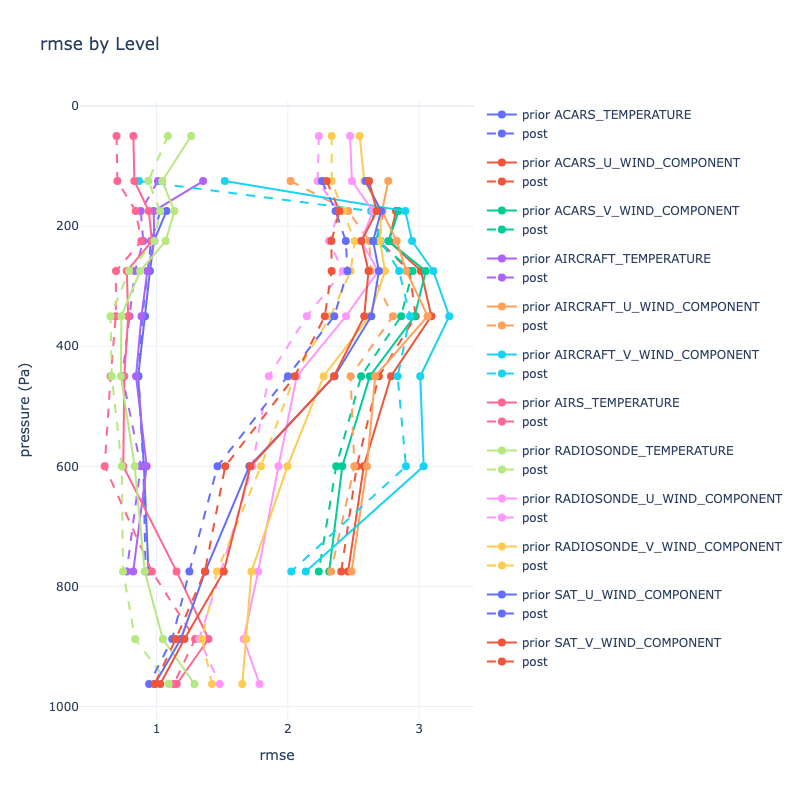
|
|
385
396
|
|
|
386
|
-

|
|
397
|
+
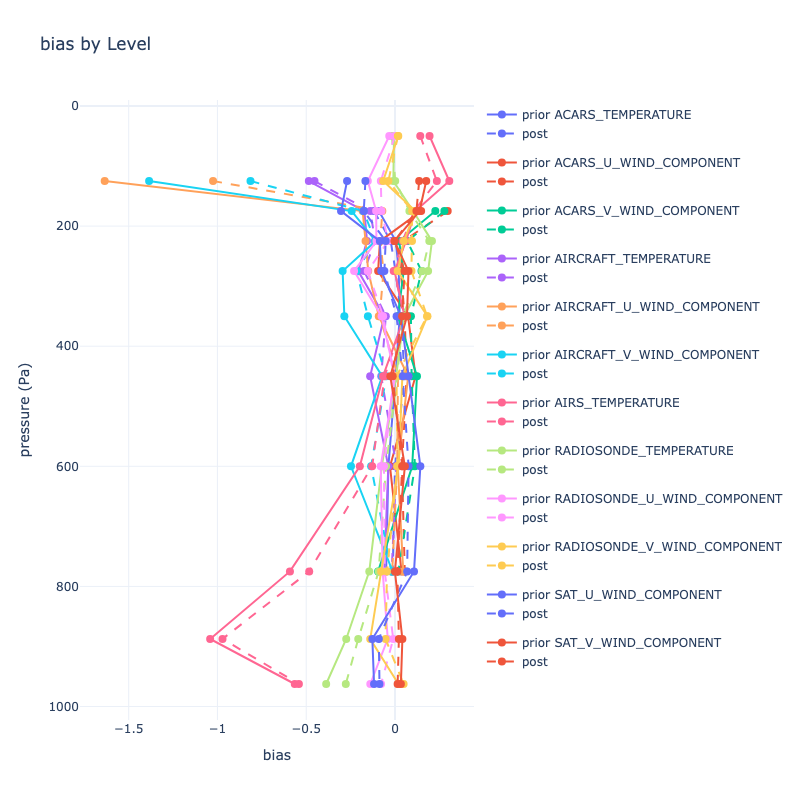
|
|
387
398
|
|
|
388
399
|
## Contributing
|
|
389
400
|
Contributions are welcome! If you have a feature request, bug report, or a suggestion, please open an issue on our GitHub repository.
|
|
390
401
|
|
|
391
402
|
## License
|
|
392
403
|
|
|
393
|
-
|
|
404
|
+
pyDARTdiags is released under the Apache License 2.0. For more details, see the LICENSE file in the root directory of this source tree or visit [Apache License 2.0](https://www.apache.org/licenses/LICENSE-2.0).
|
|
@@ -1,12 +1,20 @@
|
|
|
1
|
+
[](https://opensource.org/licenses/Apache-2.0)
|
|
2
|
+
[](https://codecov.io/gh/NCAR/pyDARTdiags)
|
|
3
|
+
[](https://pypi.org/project/pydartdiags/)
|
|
4
|
+
|
|
5
|
+
|
|
1
6
|
# pyDARTdiags
|
|
2
7
|
|
|
3
|
-
pyDARTdiags is a
|
|
8
|
+
pyDARTdiags is a Python library for obsevation space diagnostics for the Data Assimilation Research Testbed ([DART](https://github.com/NCAR/DART)).
|
|
4
9
|
|
|
5
10
|
pyDARTdiags is under initial development, so please use caution.
|
|
6
11
|
The MATLAB [observation space diagnostics](https://docs.dart.ucar.edu/en/latest/guide/matlab-observation-space.html) are available through [DART](https://github.com/NCAR/DART).
|
|
7
12
|
|
|
8
13
|
|
|
9
|
-
pyDARTdiags can be installed through pip.
|
|
14
|
+
pyDARTdiags can be installed through pip: https://pypi.org/project/pydartdiags/
|
|
15
|
+
Documenation : https://ncar.github.io/pyDARTdiags/
|
|
16
|
+
|
|
17
|
+
We recommend installing pydartdiags in a virtual enviroment:
|
|
10
18
|
|
|
11
19
|
|
|
12
20
|
```
|
|
@@ -18,14 +26,14 @@ pip install pydartdiags
|
|
|
18
26
|
## Example importing the obs\_sequence and plots modules
|
|
19
27
|
|
|
20
28
|
```python
|
|
21
|
-
from pydartdiags.obs_sequence import obs_sequence as
|
|
29
|
+
from pydartdiags.obs_sequence import obs_sequence as obsq
|
|
22
30
|
from pydartdiags.plots import plots
|
|
23
31
|
```
|
|
24
32
|
|
|
25
33
|
## Examining the dataframe
|
|
26
34
|
|
|
27
35
|
```python
|
|
28
|
-
obs_seq =
|
|
36
|
+
obs_seq = obsq.obs_sequence('obs_seq.final.ascii')
|
|
29
37
|
obs_seq.df.head()
|
|
30
38
|
```
|
|
31
39
|
|
|
@@ -186,7 +194,7 @@ obs_seq.df.head()
|
|
|
186
194
|
Find the numeber of assimilated (used) observations vs. possible observations by type
|
|
187
195
|
|
|
188
196
|
```python
|
|
189
|
-
|
|
197
|
+
obsq.possible_vs_used(obs_seq.df)
|
|
190
198
|
```
|
|
191
199
|
|
|
192
200
|
<table border="1" class="dataframe">
|
|
@@ -343,10 +351,10 @@ obs_seq.possible_vs_used(obs_seq.df)
|
|
|
343
351
|
* plot the rank histogram
|
|
344
352
|
|
|
345
353
|
```python
|
|
346
|
-
df_qc0 =
|
|
354
|
+
df_qc0 = obsq.select_by_dart_qc(obs_seq.df, 0)
|
|
347
355
|
plots.plot_rank_histogram(df_qc0)
|
|
348
356
|
```
|
|
349
|
-

|
|
357
|
+

|
|
350
358
|
|
|
351
359
|
|
|
352
360
|
### plot profile of RMSE and Bias
|
|
@@ -359,17 +367,17 @@ plots.plot_rank_histogram(df_qc0)
|
|
|
359
367
|
hPalevels = [0.0, 100.0, 150.0, 200.0, 250.0, 300.0, 400.0, 500.0, 700, 850, 925, 1000]# float("inf")] # Pa?
|
|
360
368
|
plevels = [i * 100 for i in hPalevels]
|
|
361
369
|
|
|
362
|
-
df_qc0 =
|
|
370
|
+
df_qc0 = obsq.select_by_dart_qc(obs_seq.df, 0) # only qc 0
|
|
363
371
|
df_profile, figrmse, figbias = plots.plot_profile(df_qc0, plevels)
|
|
364
372
|
```
|
|
365
373
|
|
|
366
|
-

|
|
374
|
+

|
|
367
375
|
|
|
368
|
-

|
|
376
|
+

|
|
369
377
|
|
|
370
378
|
## Contributing
|
|
371
379
|
Contributions are welcome! If you have a feature request, bug report, or a suggestion, please open an issue on our GitHub repository.
|
|
372
380
|
|
|
373
381
|
## License
|
|
374
382
|
|
|
375
|
-
|
|
383
|
+
pyDARTdiags is released under the Apache License 2.0. For more details, see the LICENSE file in the root directory of this source tree or visit [Apache License 2.0](https://www.apache.org/licenses/LICENSE-2.0).
|
|
@@ -1,10 +1,10 @@
|
|
|
1
1
|
[build-system]
|
|
2
|
-
requires = ["
|
|
3
|
-
build-backend = "
|
|
2
|
+
requires = ["setuptools", "wheel"]
|
|
3
|
+
build-backend = "setuptools.build_meta"
|
|
4
4
|
|
|
5
5
|
[project]
|
|
6
6
|
name = "pydartdiags"
|
|
7
|
-
version = "0.0.
|
|
7
|
+
version = "0.0.42"
|
|
8
8
|
authors = [
|
|
9
9
|
{ name="Helen Kershaw", email="hkershaw@ucar.edu" },
|
|
10
10
|
]
|
|
@@ -19,7 +19,8 @@ classifiers = [
|
|
|
19
19
|
dependencies = [
|
|
20
20
|
"pandas>=2.2.0",
|
|
21
21
|
"numpy>=1.26",
|
|
22
|
-
"plotly>=5.22.0"
|
|
22
|
+
"plotly>=5.22.0",
|
|
23
|
+
"pyyaml>=6.0.2"
|
|
23
24
|
]
|
|
24
25
|
|
|
25
26
|
[project.urls]
|
|
@@ -27,5 +28,3 @@ Homepage = "https://github.com/NCAR/pyDARTdiags.git"
|
|
|
27
28
|
Issues = "https://github.com/NCAR/pyDARTdiags/issues"
|
|
28
29
|
Documentation = "https://ncar.github.io/pyDARTdiags"
|
|
29
30
|
|
|
30
|
-
[tool.hatch.build.targets.wheel]
|
|
31
|
-
packages = ["src/pydartdiags"]
|
|
@@ -0,0 +1,26 @@
|
|
|
1
|
+
from setuptools import setup, find_packages
|
|
2
|
+
|
|
3
|
+
setup(
|
|
4
|
+
name="pydartdiags",
|
|
5
|
+
version="0.0.41",
|
|
6
|
+
packages=find_packages(where="src"),
|
|
7
|
+
package_dir={"": "src"},
|
|
8
|
+
author="Helen Kershaw",
|
|
9
|
+
author_email="hkershaw@ucar.edu",
|
|
10
|
+
description="Observation Sequence Diagnostics for DART",
|
|
11
|
+
long_description=open("README.md").read(),
|
|
12
|
+
long_description_content_type="text/markdown",
|
|
13
|
+
url="https://github.com/NCAR/pyDARTdiags.git",
|
|
14
|
+
classifiers=[
|
|
15
|
+
"Programming Language :: Python :: 3",
|
|
16
|
+
"License :: OSI Approved :: Apache Software License",
|
|
17
|
+
"Operating System :: OS Independent",
|
|
18
|
+
],
|
|
19
|
+
python_requires=">=3.8",
|
|
20
|
+
install_requires=[
|
|
21
|
+
"pandas>=2.2.0",
|
|
22
|
+
"numpy>=1.26",
|
|
23
|
+
"plotly>=5.22.0",
|
|
24
|
+
"pyyaml>=6.0.2"
|
|
25
|
+
],
|
|
26
|
+
)
|
|
@@ -3,6 +3,7 @@ import datetime as dt
|
|
|
3
3
|
import numpy as np
|
|
4
4
|
import os
|
|
5
5
|
import yaml
|
|
6
|
+
import struct
|
|
6
7
|
|
|
7
8
|
class obs_sequence:
|
|
8
9
|
"""Create an obs_sequence object from an ascii observation sequence file.
|
|
@@ -55,21 +56,40 @@ class obs_sequence:
|
|
|
55
56
|
|
|
56
57
|
reversed_vert = {value: key for key, value in vert.items()}
|
|
57
58
|
|
|
58
|
-
# synonyms for observation
|
|
59
|
-
synonyms_for_obs = ['NCEP BUFR observation',
|
|
60
|
-
'AIRS observation',
|
|
61
|
-
'GTSPP observation',
|
|
62
|
-
'SST observation',
|
|
63
|
-
'observations']
|
|
64
59
|
|
|
65
|
-
def __init__(self, file):
|
|
60
|
+
def __init__(self, file, synonyms=None):
|
|
66
61
|
self.loc_mod = 'None'
|
|
67
62
|
self.file = file
|
|
68
|
-
self.
|
|
63
|
+
self.synonyms_for_obs = ['NCEP BUFR observation',
|
|
64
|
+
'AIRS observation',
|
|
65
|
+
'GTSPP observation',
|
|
66
|
+
'SST observation',
|
|
67
|
+
'observations',
|
|
68
|
+
'WOD observation']
|
|
69
|
+
if synonyms:
|
|
70
|
+
if isinstance(synonyms, list):
|
|
71
|
+
self.synonyms_for_obs.extend(synonyms)
|
|
72
|
+
else:
|
|
73
|
+
self.synonyms_for_obs.append(synonyms)
|
|
74
|
+
|
|
75
|
+
module_dir = os.path.dirname(__file__)
|
|
76
|
+
self.default_composite_types = os.path.join(module_dir,"composite_types.yaml")
|
|
77
|
+
|
|
78
|
+
if self.is_binary(file):
|
|
79
|
+
self.header = self.read_binary_header(file)
|
|
80
|
+
else:
|
|
81
|
+
self.header = self.read_header(file)
|
|
82
|
+
|
|
69
83
|
self.types = self.collect_obs_types(self.header)
|
|
70
84
|
self.reverse_types = {v: k for k, v in self.types.items()}
|
|
71
85
|
self.copie_names, self.n_copies = self.collect_copie_names(self.header)
|
|
72
|
-
|
|
86
|
+
|
|
87
|
+
if self.is_binary(file):
|
|
88
|
+
self.seq = self.obs_binary_reader(file, self.n_copies)
|
|
89
|
+
self.loc_mod = 'loc3d' # only loc3d supported for binary, & no way to check
|
|
90
|
+
else:
|
|
91
|
+
self.seq = self.obs_reader(file, self.n_copies)
|
|
92
|
+
|
|
73
93
|
self.all_obs = self.create_all_obs() # uses up the generator
|
|
74
94
|
# at this point you know if the seq is loc3d or loc1d
|
|
75
95
|
if self.loc_mod == 'None':
|
|
@@ -87,8 +107,7 @@ class obs_sequence:
|
|
|
87
107
|
if 'prior_ensemble_mean'.casefold() in map(str.casefold, self.columns):
|
|
88
108
|
self.df['bias'] = (self.df['prior_ensemble_mean'] - self.df['observation'])
|
|
89
109
|
self.df['sq_err'] = self.df['bias']**2 # squared error
|
|
90
|
-
|
|
91
|
-
self.default_composite_types = os.path.join(module_dir,"composite_types.yaml")
|
|
110
|
+
|
|
92
111
|
|
|
93
112
|
def create_all_obs(self):
|
|
94
113
|
""" steps through the generator to create a
|
|
@@ -127,7 +146,11 @@ class obs_sequence:
|
|
|
127
146
|
raise ValueError("Neither 'loc3d' nor 'loc1d' could be found in the observation sequence.")
|
|
128
147
|
typeI = obs.index('kind') # type of observation
|
|
129
148
|
type_value = obs[typeI + 1]
|
|
130
|
-
|
|
149
|
+
if not self.types:
|
|
150
|
+
data.append('Identity')
|
|
151
|
+
else:
|
|
152
|
+
data.append(self.types[type_value]) # observation type
|
|
153
|
+
|
|
131
154
|
# any observation specific obs def info is between here and the end of the list
|
|
132
155
|
time = obs[-2].split()
|
|
133
156
|
data.append(int(time[0])) # seconds
|
|
@@ -256,9 +279,19 @@ class obs_sequence:
|
|
|
256
279
|
heading.append('obs_err_var')
|
|
257
280
|
return heading
|
|
258
281
|
|
|
282
|
+
@staticmethod
|
|
283
|
+
def is_binary(file):
|
|
284
|
+
"""Check if a file is binary file."""
|
|
285
|
+
with open(file, 'rb') as f:
|
|
286
|
+
chunk = f.read(1024)
|
|
287
|
+
if b'\0' in chunk:
|
|
288
|
+
return True
|
|
289
|
+
return False
|
|
290
|
+
|
|
291
|
+
|
|
259
292
|
@staticmethod
|
|
260
293
|
def read_header(file):
|
|
261
|
-
"""Read the header and number of lines in the header of an obs_seq file"""
|
|
294
|
+
"""Read the header and number of lines in the header of an ascii obs_seq file"""
|
|
262
295
|
header = []
|
|
263
296
|
with open(file, 'r') as f:
|
|
264
297
|
for line in f:
|
|
@@ -269,6 +302,118 @@ class obs_sequence:
|
|
|
269
302
|
header.append(line.strip())
|
|
270
303
|
return header
|
|
271
304
|
|
|
305
|
+
@staticmethod
|
|
306
|
+
def read_binary_header(file):
|
|
307
|
+
"""Read the header and number of lines in the header of a binary obs_seq file from Fortran output"""
|
|
308
|
+
header = []
|
|
309
|
+
linecount = 0
|
|
310
|
+
obs_types_definitions = -1000
|
|
311
|
+
num_obs = 0
|
|
312
|
+
max_num_obs = 0
|
|
313
|
+
# need to get:
|
|
314
|
+
# number of obs_type_definitions
|
|
315
|
+
# number of copies
|
|
316
|
+
# number of qcs
|
|
317
|
+
with open(file, 'rb') as f:
|
|
318
|
+
while True:
|
|
319
|
+
# Read the record length
|
|
320
|
+
record_length = obs_sequence.read_record_length(f)
|
|
321
|
+
if record_length is None:
|
|
322
|
+
break
|
|
323
|
+
record = f.read(record_length)
|
|
324
|
+
if not record: # end of file
|
|
325
|
+
break
|
|
326
|
+
|
|
327
|
+
# Read the trailing record length (should match the leading one)
|
|
328
|
+
obs_sequence.check_trailing_record_length(f, record_length)
|
|
329
|
+
|
|
330
|
+
linecount += 1
|
|
331
|
+
|
|
332
|
+
if linecount == 3:
|
|
333
|
+
obs_types_definitions = struct.unpack('i', record)[0]
|
|
334
|
+
continue
|
|
335
|
+
|
|
336
|
+
if linecount == 4+obs_types_definitions:
|
|
337
|
+
num_copies, num_qcs, num_obs, max_num_obs = struct.unpack('iiii', record)[:16]
|
|
338
|
+
break
|
|
339
|
+
|
|
340
|
+
# Go back to the beginning of the file
|
|
341
|
+
f.seek(0)
|
|
342
|
+
|
|
343
|
+
for _ in range(2):
|
|
344
|
+
record_length = obs_sequence.read_record_length(f)
|
|
345
|
+
if record_length is None:
|
|
346
|
+
break
|
|
347
|
+
|
|
348
|
+
record = f.read(record_length)
|
|
349
|
+
if not record: # end of file
|
|
350
|
+
break
|
|
351
|
+
|
|
352
|
+
obs_sequence.check_trailing_record_length(f, record_length)
|
|
353
|
+
header.append(record.decode('utf-8').strip())
|
|
354
|
+
|
|
355
|
+
header.append(str(obs_types_definitions))
|
|
356
|
+
|
|
357
|
+
# obs_types_definitions
|
|
358
|
+
for _ in range(3,4+obs_types_definitions):
|
|
359
|
+
# Read the record length
|
|
360
|
+
record_length = obs_sequence.read_record_length(f)
|
|
361
|
+
if record_length is None:
|
|
362
|
+
break
|
|
363
|
+
|
|
364
|
+
# Read the actual record
|
|
365
|
+
record = f.read(record_length)
|
|
366
|
+
if not record: # end of file
|
|
367
|
+
break
|
|
368
|
+
|
|
369
|
+
obs_sequence.check_trailing_record_length(f, record_length)
|
|
370
|
+
|
|
371
|
+
if _ == 3:
|
|
372
|
+
continue # num obs_types_definitions
|
|
373
|
+
# Read an integer and a string from the record
|
|
374
|
+
integer_value = struct.unpack('i', record[:4])[0]
|
|
375
|
+
string_value = record[4:].decode('utf-8').strip()
|
|
376
|
+
header.append(f"{integer_value} {string_value}")
|
|
377
|
+
|
|
378
|
+
header.append(f"num_copies: {num_copies} num_qc: {num_qcs}")
|
|
379
|
+
header.append(f"num_obs: {num_obs} max_num_obs: {max_num_obs}")
|
|
380
|
+
|
|
381
|
+
#copie names
|
|
382
|
+
for _ in range(5+obs_types_definitions, 5+obs_types_definitions+num_copies+num_qcs+1):
|
|
383
|
+
# Read the record length
|
|
384
|
+
record_length = obs_sequence.read_record_length(f)
|
|
385
|
+
if record_length is None:
|
|
386
|
+
break
|
|
387
|
+
|
|
388
|
+
# Read the actual record
|
|
389
|
+
record = f.read(record_length)
|
|
390
|
+
if not record:
|
|
391
|
+
break
|
|
392
|
+
|
|
393
|
+
obs_sequence.check_trailing_record_length(f, record_length)
|
|
394
|
+
|
|
395
|
+
if _ == 5+obs_types_definitions:
|
|
396
|
+
continue
|
|
397
|
+
|
|
398
|
+
# Read the whole record as a string
|
|
399
|
+
string_value = record.decode('utf-8').strip()
|
|
400
|
+
header.append(string_value)
|
|
401
|
+
|
|
402
|
+
# first and last obs
|
|
403
|
+
# Read the record length
|
|
404
|
+
record_length = obs_sequence.read_record_length(f)
|
|
405
|
+
|
|
406
|
+
# Read the actual record
|
|
407
|
+
record = f.read(record_length)
|
|
408
|
+
|
|
409
|
+
obs_sequence.check_trailing_record_length(f, record_length)
|
|
410
|
+
|
|
411
|
+
# Read the whole record as a two integers
|
|
412
|
+
first, last = struct.unpack('ii', record)[:8]
|
|
413
|
+
header.append(f"first: {first} last: {last}")
|
|
414
|
+
|
|
415
|
+
return header
|
|
416
|
+
|
|
272
417
|
@staticmethod
|
|
273
418
|
def collect_obs_types(header):
|
|
274
419
|
"""Create a dictionary for the observation types in the obs_seq header"""
|
|
@@ -298,7 +443,7 @@ class obs_sequence:
|
|
|
298
443
|
|
|
299
444
|
@staticmethod
|
|
300
445
|
def obs_reader(file, n):
|
|
301
|
-
"""Reads the obs sequence file and returns a generator of the obs"""
|
|
446
|
+
"""Reads the ascii obs sequence file and returns a generator of the obs"""
|
|
302
447
|
previous_line = ''
|
|
303
448
|
with open(file, 'r') as f:
|
|
304
449
|
for line in f:
|
|
@@ -338,6 +483,115 @@ class obs_sequence:
|
|
|
338
483
|
previous_line = next_line
|
|
339
484
|
yield obs
|
|
340
485
|
|
|
486
|
+
@staticmethod
|
|
487
|
+
def check_trailing_record_length(file, expected_length):
|
|
488
|
+
"""Reads and checks the trailing record length from the binary file written by Fortran.
|
|
489
|
+
|
|
490
|
+
Parameters:
|
|
491
|
+
file (file): The file object.
|
|
492
|
+
expected_length (int): The expected length of the trailing record.
|
|
493
|
+
|
|
494
|
+
Assuming 4 bytes:
|
|
495
|
+
| Record Length (4 bytes) | Data (N bytes) | Trailing Record Length (4 bytes) |
|
|
496
|
+
"""
|
|
497
|
+
trailing_record_length_bytes = file.read(4)
|
|
498
|
+
trailing_record_length = struct.unpack('i', trailing_record_length_bytes)[0]
|
|
499
|
+
if expected_length != trailing_record_length:
|
|
500
|
+
raise ValueError("Record length mismatch in Fortran binary file")
|
|
501
|
+
|
|
502
|
+
@staticmethod
|
|
503
|
+
def read_record_length(file):
|
|
504
|
+
"""Reads and unpacks the record length from the file."""
|
|
505
|
+
record_length_bytes = file.read(4)
|
|
506
|
+
if not record_length_bytes:
|
|
507
|
+
return None # End of file
|
|
508
|
+
return struct.unpack('i', record_length_bytes)[0]
|
|
509
|
+
|
|
510
|
+
|
|
511
|
+
def obs_binary_reader(self, file, n):
|
|
512
|
+
"""Reads the obs sequence binary file and returns a generator of the obs"""
|
|
513
|
+
header_length = len(self.header)
|
|
514
|
+
with open(file, 'rb') as f:
|
|
515
|
+
# Skip the first len(obs_seq.header) lines
|
|
516
|
+
for _ in range(header_length-1):
|
|
517
|
+
# Read the record length
|
|
518
|
+
record_length = obs_sequence.read_record_length(f)
|
|
519
|
+
if record_length is None: # End of file
|
|
520
|
+
break
|
|
521
|
+
|
|
522
|
+
# Skip the actual record
|
|
523
|
+
f.seek(record_length, 1)
|
|
524
|
+
|
|
525
|
+
# Skip the trailing record length
|
|
526
|
+
f.seek(4, 1)
|
|
527
|
+
|
|
528
|
+
obs_num = 0
|
|
529
|
+
while True:
|
|
530
|
+
obs = []
|
|
531
|
+
obs_num += 1
|
|
532
|
+
obs.append(f"OBS {obs_num}")
|
|
533
|
+
for _ in range(n): # number of copies
|
|
534
|
+
# Read the record length
|
|
535
|
+
record_length = obs_sequence.read_record_length(f)
|
|
536
|
+
if record_length is None:
|
|
537
|
+
break
|
|
538
|
+
# Read the actual record (copie)
|
|
539
|
+
record = f.read(record_length)
|
|
540
|
+
obs.append(struct.unpack('d', record)[0])
|
|
541
|
+
|
|
542
|
+
# Read the trailing record length (should match the leading one)
|
|
543
|
+
obs_sequence.check_trailing_record_length(f, record_length)
|
|
544
|
+
|
|
545
|
+
# linked list info
|
|
546
|
+
record_length = obs_sequence.read_record_length(f)
|
|
547
|
+
if record_length is None:
|
|
548
|
+
break
|
|
549
|
+
|
|
550
|
+
record = f.read(record_length)
|
|
551
|
+
int1, int2, int3 = struct.unpack('iii', record[:12])
|
|
552
|
+
linked_list_string = f"{int1:<12} {int2:<10} {int3:<12}"
|
|
553
|
+
obs.append(linked_list_string)
|
|
554
|
+
|
|
555
|
+
obs_sequence.check_trailing_record_length(f, record_length)
|
|
556
|
+
|
|
557
|
+
# location (note no location header "loc3d" or "loc1d" for binary files)
|
|
558
|
+
obs.append('loc3d')
|
|
559
|
+
record_length = obs_sequence.read_record_length(f)
|
|
560
|
+
record = f.read(record_length)
|
|
561
|
+
x,y,z,vert = struct.unpack('dddi', record[:28])
|
|
562
|
+
location_string = f"{x} {y} {z} {vert}"
|
|
563
|
+
obs.append(location_string)
|
|
564
|
+
|
|
565
|
+
obs_sequence.check_trailing_record_length(f, record_length)
|
|
566
|
+
|
|
567
|
+
# kind (type of observation) value
|
|
568
|
+
obs.append('kind')
|
|
569
|
+
record_length_bytes = f.read(4)
|
|
570
|
+
record_length = struct.unpack('i', record_length_bytes)[0]
|
|
571
|
+
record = f.read(record_length)
|
|
572
|
+
kind = f"{struct.unpack('i', record)[0]}"
|
|
573
|
+
obs.append(kind)
|
|
574
|
+
|
|
575
|
+
obs_sequence.check_trailing_record_length(f, record_length)
|
|
576
|
+
|
|
577
|
+
# time (seconds, days)
|
|
578
|
+
record_length = obs_sequence.read_record_length(f)
|
|
579
|
+
record = f.read(record_length)
|
|
580
|
+
seconds, days = struct.unpack('ii', record)[:8]
|
|
581
|
+
time_string = f"{seconds} {days}"
|
|
582
|
+
obs.append(time_string)
|
|
583
|
+
|
|
584
|
+
obs_sequence.check_trailing_record_length(f, record_length)
|
|
585
|
+
|
|
586
|
+
# obs error variance
|
|
587
|
+
record_length = obs_sequence.read_record_length(f)
|
|
588
|
+
record = f.read(record_length)
|
|
589
|
+
obs.append(struct.unpack('d', record)[0])
|
|
590
|
+
|
|
591
|
+
obs_sequence.check_trailing_record_length(f, record_length)
|
|
592
|
+
|
|
593
|
+
yield obs
|
|
594
|
+
|
|
341
595
|
def composite_types(self, composite_types='use_default'):
|
|
342
596
|
"""
|
|
343
597
|
Set up and construct composite types for the DataFrame.
|
|
@@ -503,3 +757,5 @@ def construct_composit(df_comp, composite, components):
|
|
|
503
757
|
merged_df = merged_df.drop(columns=[col for col in merged_df.columns if col.endswith('_v')])
|
|
504
758
|
|
|
505
759
|
return merged_df
|
|
760
|
+
|
|
761
|
+
|
|
@@ -29,7 +29,7 @@ def calculate_rank(df):
|
|
|
29
29
|
|
|
30
30
|
Parameters:
|
|
31
31
|
df (pd.DataFrame): A DataFrame with columns for mean, standard deviation, observed values,
|
|
32
|
-
|
|
32
|
+
ensemble size, and observation type. The DataFrame should have one row per observation.
|
|
33
33
|
|
|
34
34
|
Returns:
|
|
35
35
|
tuple: A tuple containing the rank array, ensemble size, and a result DataFrame. The result
|
|
@@ -125,7 +125,9 @@ def mean_then_sqrt(x):
|
|
|
125
125
|
|
|
126
126
|
Raises:
|
|
127
127
|
TypeError: If the input is not an array-like object containing numeric values.
|
|
128
|
+
ValueError: If the input array is empty.
|
|
128
129
|
"""
|
|
130
|
+
|
|
129
131
|
return np.sqrt(np.mean(x))
|
|
130
132
|
|
|
131
133
|
def rmse_bias(df):
|